Immuno-Dipstick for Colletotrichum gloeosporioides Detection: Towards On-Farm Application
- PMID: 35200310
- PMCID: PMC8869205
- DOI: 10.3390/bios12020049
Immuno-Dipstick for Colletotrichum gloeosporioides Detection: Towards On-Farm Application
Abstract
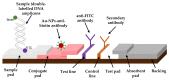
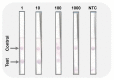
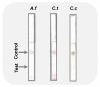
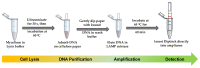
Early and quick detection of pathogens are crucial for managing the spread of infections in the biomedical, biosafety, food, and agricultural fields. While molecular diagnostics can offer the specificity and reliability in acute infectious diseases, detection of pathogens is often slowed down by the current benchtop molecular diagnoses, which are time consuming, labor intensive, and lack the mobility for application at the point-of-need. In this work, we developed a complete on-farm use detection protocol for the plant-devastating anthracnose agent: Colletotrichum gloeosporioides. Our methods combined a simplified DNA extraction on paper that is compatible with loop-mediated isothermal amplification (LAMP), coupled with paper-based immunoassay lateral flow sensing. Our results offer simple, quick, easy, and a minimally instrumented toolkit for Colletotrichum gloeosporioides detection. This scalable and adaptable platform is a valuable alternative to traditional sensing systems towards on-the-go pathogen detection in food and agriculture, biomedical, and other fields.
Keywords: Colletotrichum gloeosporioides; anthracnose; immuno-dipstick; loop-mediated amplification (LAMP); point-of-need molecular diagnostic; simplified DNA extraction.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Gupta V., Pandey A., Kumar P., Pandey B., Gaur R., Bajpai V., Sharma N., Sharma S. Genetic Characterization of Mango Anthracnose Pathogen Colletotrichum gloeosporioides Penz. by Random Amplified Polymorphic DNA Analysis. Afr. J. Biotechnol. 2010;9:4009–4013.
-
- Kelly J.D., Vallejo V.A. A Comprehensive Review of the Major Genes Conditioning Resistance to Anthracnose in Common Bean. HortScience. 2004;39:1196–1207. doi: 10.21273/HORTSCI.39.6.1196. - DOI
-
- Heydari A., Pessarakli M. A Review on Biological Control of Fungal Plant Pathogens Using Microbial Antagonists. J. Biol. Sci. 2010;10:273–290. doi: 10.3923/jbs.2010.273.290. - DOI
-
- Min J.-S., Kim K.-S., Kim S.-W., Jung J.-H., Lamsal K., Kim S.-B., Jung M.-Y., Lee Y.-S. Effects of Colloidal Silver Nanoparticles on Sclerotium-Forming Phytopathogenic Fungi. Plant Pathol. J. 2009;25:376–380. doi: 10.5423/PPJ.2009.25.4.376. - DOI
MeSH terms
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources

