A FRET-Based Biosensor for the Src N-Terminal Regulatory Element
- PMID: 35200356
- PMCID: PMC8870054
- DOI: 10.3390/bios12020096
A FRET-Based Biosensor for the Src N-Terminal Regulatory Element
Abstract
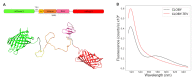
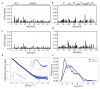
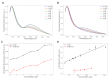
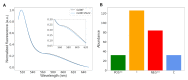
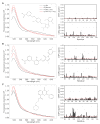
In signaling proteins, intrinsically disordered regions often represent regulatory elements, which are sensitive to environmental effects, ligand binding, and post-translational modifications. The conformational space sampled by disordered regions can be affected by environmental stimuli and these changes trigger, vis a vis effector domain, downstream processes. The disordered nature of these regulatory elements enables signal integration and graded responses but prevents the application of classical approaches for drug screening based on the existence of a fixed three-dimensional structure. We have designed a genetically encodable biosensor for the N-terminal regulatory element of the c-Src kinase, the first discovered protooncogene and lead representative of the Src family of kinases. The biosensor is formed by two fluorescent proteins forming a FRET pair fused at the two extremes of a construct including the SH4, unique and SH3 domains of Src. An internal control is provided by an engineered proteolytic site allowing the generation of an identical mixture of the disconnected fluorophores. We show FRET variations induced by ligand binding. The biosensor has been used for a high-throughput screening of a library of 1669 compounds with seven hits confirmed by NMR.
Keywords: NMR; c-Src; fluorescence; fuzzy complexes; high-throughput screening; intrinsically disordered proteins (IDP).
Conflict of interest statement
IDP Discovery Pharma is a company developing drugs for disordered proteins. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures
References
-
- Uversky V.N. Intrinsically disordered proteins and their ‘Mysterious’ (meta)physics. Front. Phys. 2019;7:10. doi: 10.3389/fphy.2019.00010. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

