Quantitative Studies on the Interaction between Saposin-like Proteins and Synthetic Lipid Membranes
- PMID: 35200535
- PMCID: PMC8878781
- DOI: 10.3390/mps5010019
Quantitative Studies on the Interaction between Saposin-like Proteins and Synthetic Lipid Membranes
Abstract
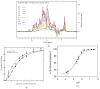
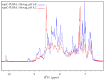
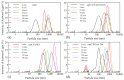
Members of the saposin-fold protein family and related proteins sharing a similar fold (saposin-like proteins; SAPLIP) are peripheral-membrane binding proteins that perform essential cellular functions. Saposins and SAPLIPs are abundant in both plant and animal kingdoms, and peripherally bind to lipid membranes to play important roles in lipid transfer and hydrolysis, defense mechanisms, surfactant stabilization, and cell proliferation. However, quantitative studies on the interaction between proteins and membranes are challenging due to the different nature of the two components in relation to size, structure, chemical composition, and polarity. Using liposomes and the saposin-fold member saposin C (sapC) as model systems, we describe here a method to apply solution NMR and dynamic light scattering to study the interaction between SAPLIPs and synthetic membranes at the quantitative level. Specifically, we prove with NMR that sapC binds reversibly to the synthetic membrane in a pH-controlled manner and show the dynamic nature of its fusogenic properties with dynamic light scattering. The method can be used to infer the optimal pH for membrane binding and to determine an apparent dissociation constant (KDapp) for protein-liposome interaction. We propose that these experiments can be applied to other proteins sharing the saposin fold.
Keywords: NMR; dynamic light scattering; liposomes; membrane fusion; protein-membrane interactions; saposin C; saposin-like proteins.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
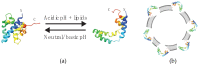
References
Grants and funding
LinkOut - more resources
Full Text Sources

