Division site determination during asymmetric cell division in plants
- PMID: 35201345
- PMCID: PMC9134084
- DOI: 10.1093/plcell/koac069
Division site determination during asymmetric cell division in plants
Abstract
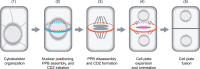
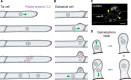
During development, both animals and plants exploit asymmetric cell division (ACD) to increase tissue complexity, a process that usually generates cells dissimilar in size, morphology, and fate. Plants lack the key regulators that control ACD in animals. Instead, plants have evolved two unique cytoskeletal structures to tackle this problem: the preprophase band (PPB) and phragmoplast. The assembly of the PPB and phragmoplast and their contributions to division plane orientation have been extensively studied. However, how the division plane is positioned off the cell center during asymmetric division is poorly understood. Over the past 20 years, emerging evidence points to a critical role for polarly localized membrane proteins in this process. Although many of these proteins are species- or cell type specific, and the molecular mechanism underlying division asymmetry is not fully understood, common features such as morphological changes in cells, cytoskeletal dynamics, and nuclear positioning have been observed. In this review, we provide updates on polarity establishment and nuclear positioning during ACD in plants. Together with previous findings about symmetrically dividing cells and the emerging roles of developmental cues, we aim to offer evolutionary insight into a common framework for asymmetric division-site determination and highlight directions for future work.
© American Society of Plant Biologists 2022. All rights reserved. For permissions, please email: journals.permissions@oup.com.
Figures




Similar articles
-
The cytoplast concept in dividing plant cells: cytoplasmic domains and the evolution of spatially organized cell.Am J Bot. 1999 Feb;86(2):153-72. Am J Bot. 1999. PMID: 21680355
-
A polarized nuclear position specifies the correct division plane during maize stomatal development.Plant Physiol. 2023 Aug 31;193(1):125-139. doi: 10.1093/plphys/kiad329. Plant Physiol. 2023. PMID: 37300534
-
Determination of symmetric and asymmetric division planes in plant cells.Annu Rev Plant Biol. 2011;62:387-409. doi: 10.1146/annurev-arplant-042110-103802. Annu Rev Plant Biol. 2011. PMID: 21391814 Review.
-
Spindle motility skews division site determination during asymmetric cell division in Physcomitrella.Nat Commun. 2022 May 5;13(1):2488. doi: 10.1038/s41467-022-30239-1. Nat Commun. 2022. PMID: 35513464 Free PMC article.
-
The role of the cytoskeleton and associated proteins in determination of the plant cell division plane.Plant J. 2013 Jul;75(2):258-69. doi: 10.1111/tpj.12177. Epub 2013 Apr 19. Plant J. 2013. PMID: 23496276 Review.
Cited by
-
Rab GTPases, tethers, and SNAREs work together to regulate Arabidopsis cell plate formation.Front Plant Sci. 2023 Feb 10;14:1120841. doi: 10.3389/fpls.2023.1120841. eCollection 2023. Front Plant Sci. 2023. PMID: 36844074 Free PMC article. Review.
-
RHO OF PLANTS signalling and the activating ROP GUANINE NUCLEOTIDE EXCHANGE FACTORS: specificity in cellular signal transduction in plants.J Exp Bot. 2024 Jun 24;75(12):3685-3699. doi: 10.1093/jxb/erae196. J Exp Bot. 2024. PMID: 38683617 Free PMC article. Review.
-
Is the nucleus the unwitting architect of asymmetric cell division in plants?Plant Physiol. 2023 Aug 31;193(1):6-8. doi: 10.1093/plphys/kiad379. Plant Physiol. 2023. PMID: 37399225 Free PMC article. No abstract available.
-
Asymmetric cell division in plant development.J Integr Plant Biol. 2023 Feb;65(2):343-370. doi: 10.1111/jipb.13446. J Integr Plant Biol. 2023. PMID: 36610013 Free PMC article. Review.
-
Comprehensive and quantitative analysis of intracellular structure polarization at the apical-basal axis in elongating Arabidopsis zygotes.Sci Rep. 2023 Dec 18;13(1):22879. doi: 10.1038/s41598-023-50020-8. Sci Rep. 2023. PMID: 38129559 Free PMC article.
References
-
- Abrash EB, Bergmann DC (2009) Asymmetric cell divisions: a view from plant development. Dev Cell 16: 783–796 - PubMed
-
- Ambrose JC, Cyr R (2008) Mitotic spindle organization by the preprophase band. Mol Plant 1: 950–960 - PubMed
-
- Armenta-Medina A, Gillmor CS (2019) Genetic, molecular and parent-of-origin regulation of early embryogenesis in flowering plants. Curr Top Dev Biol 131: 497–543 - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

