Deep analysis of neuroblastoma core regulatory circuitries using online databases and integrated bioinformatics shows their pan-cancer roles as prognostic predictors
- PMID: 35201514
- PMCID: PMC8777518
- DOI: 10.1007/s12672-021-00452-3
Deep analysis of neuroblastoma core regulatory circuitries using online databases and integrated bioinformatics shows their pan-cancer roles as prognostic predictors
Abstract
Aim: Neuroblastoma is a heterogeneous childhood cancer derived from the neural crest. The dual cell identities of neuroblastoma include Mesenchymal (MES) and Adrenergic (ADRN). These identities are conferred by a small set of tightly-regulated transcription factors (TFs) binding super enhancers, collectively forming core regulatory circuitries (CRCs). The purpose of this study was to gain a deep understanding of the role of MES and ADRN TFs in neuroblastoma and other cancers as potential indicators of disease prognosis, progression, and relapse.
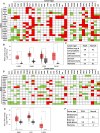
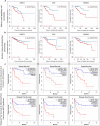
Methods: To that end, we first investigated the expression and mutational profile of MES and ADRN TFs in neuroblastoma. Moreover, we established their correlation with neuroblastoma risk groups and overall survival while establishing their extended networks with long non-coding RNAs (lncRNAs). Furthermore, we analysed the pan-cancer expression and mutational profile of these TFs and their correlation with patient survival and finally their network connectivity, using a panel of bioinformatic tools including GEPIA2, human pathology atlas, TIMER2, Omicsnet, and Cytoscape.
Results: We show the association of multiple MES and ADRN TFs with neuroblastoma risk groups and overall survival and find significantly higher expression of various MES and ADRN TFs compared to normal tissues and their association with overall survival and disease-free survival in multiple cancers. Moreover, we report the strong correlation of the expression of these TFs with the infiltration of stromal and immune cells in the tumour microenvironment and with stemness and metastasis-related genes. Furthermore, we reveal extended pan-cancer networks comprising these TFs that influence the tumour microenvironment and metastasis and may be useful indicators of cancer prognosis and patient survival.
Conclusion: Our meta-analysis shows the significance of MES and ADRN TFs as indicators of patient prognosis and the putative utility of these TFs as potential novel biomarkers.
Keywords: Core regulatory circuitry; Differentiation; Gene networks; Neuroblastoma; Solid cancers; Tumour microenvironment.
© 2021. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




References
-
- Brodeur GM. Neuroblastoma: biological insights into a clinical enigma. Nat Rev Cancer. 2003;3:203–216. - PubMed
-
- Maris JM, Hogarty MD, Bagatell R, Cohn SL. Neuroblastoma. Lancet. 2007;369:2106–2120. - PubMed
-
- van Groningen T, Koster J, Valentijn LJ, Zwijnenburg DA, Akogul N, Hasselt NE, Broekmans M, Haneveld F, Nowakowska NE, Bras J, et al. Neuroblastoma is composed of two super-enhancer-associated differentiation states. Nat Genet. 2017;49(8):1261–1266. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
