Hsa-miR-183-5p Modulates Cell Adhesion by Repression of ITGB1 Expression in Prostate Cancer
- PMID: 35202085
- PMCID: PMC8875343
- DOI: 10.3390/ncrna8010011
Hsa-miR-183-5p Modulates Cell Adhesion by Repression of ITGB1 Expression in Prostate Cancer
Abstract
Prostate cancer is a major health problem worldwide. MiR-183 is an oncomiR and a candidate biomarker in prostate cancer, affecting various pathways responsible for disease initiation and progression. We sought to discover the most relevant processes controlled by miR-183 through an unbiased transcriptomic approach using prostate cell lines and patient tissues to identify miR-183 responsive genes and pathways. Gain of function experiments, reporter gene assays, and transcript and protein measurements were conducted to validate predicted functional effects and protein mediators. A total of 135 candidate miR-183 target genes overrepresenting cell adhesion terms were inferred from the integrated transcriptomic analysis. Cell attachment, spreading assays and focal adhesion quantification of miR-183-overexpressing cells confirmed the predicted reduction in cell adhesion. ITGB1 was validated as a major target of repression by miR-183 as well as a mediator of cell adhesion in response to miR-183. The reporter gene assay and PAR-CLIP read mapping suggest that ITGB1 may be a direct target of miR-183. The negative correlation between miR-183 and ITGB1 expression in prostate cancer cohorts supports their interaction in the clinical set. Overall, cell adhesion was uncovered as a major pathway controlled by miR-183 in prostate cancer, and ITGB1 was identified as a relevant mediator of this effect.
Keywords: AGO-PAR-CLIP; ITGB1; TCGA; cancer; focal adhesion; miR-183; microRNA; prostate.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

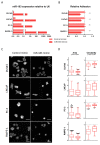
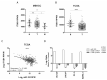


References
-
- Ambs S., Prueitt R.L., Yi M., Hudson R.S., Howe T.M., Petrocca F., Wallace T.A., Liu C.G., Volinia S., Calin G.A., et al. Genomic Profiling of MicroRNA and Messenger RNA Reveals Deregulated MicroRNA Expression in Prostate Cancer. Cancer Res. 2008;68:6162–6170. doi: 10.1158/0008-5472.CAN-08-0144. - DOI - PMC - PubMed
Grants and funding
- Research Grants 2011-Duhagon/Comisión Honoraria de Lucha contra el cáncer
- M.A.D. I+D 2016 #487/Comisión Sectorial de Investigación Científica
- M.A.D I+D 2020 # 566/Comisión Sectorial de Investigación Científica
- Annual Aliquots and Equipment Aid Programs -Duhagon/Programa para el Desarrollo de las Ciencias Básicas (PEDECIBA, MEC), Uruguay
- Annual Aliquots and Equipment Aid Programs -Duhagon/Programa para el Desarrollo de las Ciencias Básicas (PEDECIBA, MEC), Uruguay
LinkOut - more resources
Full Text Sources
Miscellaneous

