A multi-tissue study of immune gene expression profiling highlights the key role of the nasal epithelium in COVID-19 severity
- PMID: 35202626
- PMCID: PMC8861187
- DOI: 10.1016/j.envres.2022.112890
A multi-tissue study of immune gene expression profiling highlights the key role of the nasal epithelium in COVID-19 severity
Abstract
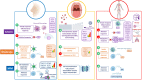
Coronavirus Disease-19 (COVID-19) symptoms range from mild to severe illness; the cause for this differential response to infection remains unknown. Unravelling the immune mechanisms acting at different levels of the colonization process might be key to understand these differences. We carried out a multi-tissue (nasal, buccal and blood; n = 156) gene expression analysis of immune-related genes from patients affected by different COVID-19 severities, and healthy controls through the nCounter technology. Mild and asymptomatic cases showed a powerful innate antiviral response in nasal epithelium, characterized by activation of interferon (IFN) pathway and downstream cascades, successfully controlling the infection at local level. In contrast, weak macrophage/monocyte driven innate antiviral response and lack of IFN signalling activity were present in severe cases. Consequently, oral mucosa from severe patients showed signals of viral activity, cell arresting and viral dissemination to the lower respiratory tract, which ultimately could explain the exacerbated innate immune response and impaired adaptative immune responses observed at systemic level. Results from saliva transcriptome suggest that the buccal cavity might play a key role in SARS-CoV-2 infection and dissemination in patients with worse prognosis. Co-expression network analysis adds further support to these findings, by detecting modules specifically correlated with severity involved in the abovementioned biological routes; this analysis also provides new candidate genes that might be tested as biomarkers in future studies. We also found tissue specific severity-related signatures mainly represented by genes involved in the innate immune system and cytokine/chemokine signalling. Local immune response could be key to determine the course of the systemic response and thus COVID-19 severity. Our findings provide a framework to investigate severity host gene biomarkers and pathways that might be relevant to diagnosis, prognosis, and therapy.
Keywords: COVID-19 severity; Co-expression analysis; Differential expression analysis; Gene expression; Immune response; Multi-tissue; Pathways analysis; SARS-CoV-2.
Copyright © 2022 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures




References
-
- Alfi O., Yakirevitch A., Wald O., Wandel O., Izhar U., Oiknine-Djian E., Nevo Y., Elgavish S., Dagan E., Madgar O., et al. Human nasal and lung tissues infected ex vivo with SARS-CoV-2 provide insights into differential tissue-specific and virus-specific innate immune responses in the upper and lower respiratory tract. J. Virol. 2021 - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

