Genomic Epidemiology of Early SARS-CoV-2 Transmission Dynamics, Gujarat, India
- PMID: 35203112
- PMCID: PMC8962880
- DOI: 10.3201/eid2804.212053
Genomic Epidemiology of Early SARS-CoV-2 Transmission Dynamics, Gujarat, India
Abstract
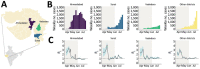
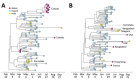
Limited genomic sampling in many high-incidence countries has impeded studies of severe respiratory syndrome coronavirus 2 (SARS-CoV-2) genomic epidemiology. Consequently, critical questions remain about the generation and global distribution of virus genetic diversity. We investigated SARS-CoV-2 transmission dynamics in Gujarat, India, during the state's first epidemic wave to shed light on spread of the virus in one of the regions hardest hit by the pandemic. By integrating case data and 434 whole-genome sequences sampled across 20 districts, we reconstructed the epidemic dynamics and spatial spread of SARS-CoV-2 in Gujarat. Our findings indicate global and regional connectivity and population density were major drivers of the Gujarat outbreak. We detected >100 virus lineage introductions, most of which appear to be associated with international travel. Within Gujarat, virus dissemination occurred predominantly from densely populated regions to geographically proximate locations that had low population density, suggesting that urban centers contributed disproportionately to virus spread.
Keywords: COVID-19; Gujarat; India; SARS-CoV-2; coronavirus disease; coronaviruses; genomic epidemiology; phylogeography; respiratory infections; severe acute respiratory syndrome coronavirus 2; transmission dynamics; virus; virus transmission; zoonoses.
Figures


References
-
- López MG, Chiner-Oms Á, García de Viedma D, Ruiz-Rodriguez P, Bracho MA, Cancino-Muñoz I, et al.; SeqCOVID-Spain consortium. The first wave of the COVID-19 epidemic in Spain was associated with early introductions and fast spread of a dominating genetic variant. Nat Genet. 2021;53:1405–14. 10.1038/s41588-021-00936-6 - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

