Epigenetic Drugs and Their Immune Modulating Potential in Cancers
- PMID: 35203421
- PMCID: PMC8868629
- DOI: 10.3390/biomedicines10020211
Epigenetic Drugs and Their Immune Modulating Potential in Cancers
Abstract
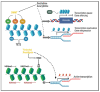
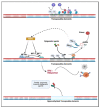
Epigenetic drugs are used for the clinical treatment of hematologic malignancies; however, their therapeutic potential in solid tumors is still under investigation. Current evidence suggests that epigenetic drugs may lead to antitumor immunity by increasing antigen presentation and may enhance the therapeutic effect of immune checkpoint inhibitors. Here, we highlight their impact on the tumor epigenome and discuss the recent evidence that epigenetic agents may optimize the immune microenvironment and promote antiviral response.
Keywords: antiviral response; epigenetic drugs; epigenetics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Epigenetic Reprogramming for Targeting IDH-Mutant Malignant Gliomas.Cancers (Basel). 2019 Oct 22;11(10):1616. doi: 10.3390/cancers11101616. Cancers (Basel). 2019. PMID: 31652645 Free PMC article. Review.
-
Clinical Potential of Kinase Inhibitors in Combination with Immune Checkpoint Inhibitors for the Treatment of Solid Tumors.Int J Mol Sci. 2021 Mar 5;22(5):2608. doi: 10.3390/ijms22052608. Int J Mol Sci. 2021. PMID: 33807608 Free PMC article. Review.
-
Cancer epigenetics and the potential of epigenetic drugs for treating solid tumors.Expert Rev Anticancer Ther. 2019 Feb;19(2):139-149. doi: 10.1080/14737140.2019.1552139. Epub 2018 Dec 4. Expert Rev Anticancer Ther. 2019. PMID: 30470148 Review.
-
Potential of epigenetic therapies in the management of solid tumors.Cancer Manag Res. 2015 Jul 31;7:241-51. doi: 10.2147/CMAR.S70358. eCollection 2015. Cancer Manag Res. 2015. PMID: 26346546 Free PMC article. Review.
-
Cancer epigenetics: new therapies and new challenges.J Drug Deliv. 2013;2013:529312. doi: 10.1155/2013/529312. Epub 2013 Feb 26. J Drug Deliv. 2013. PMID: 23533770 Free PMC article.
Cited by
-
Molecular profiling and therapeutic tailoring to address disease heterogeneity in systemic lupus erythematosus.Clin Exp Med. 2024 Sep 19;24(1):223. doi: 10.1007/s10238-024-01484-z. Clin Exp Med. 2024. PMID: 39294397 Free PMC article. Review.
-
Epigenetic marvels: exploring the landscape of colorectal cancer treatment through cutting-edge epigenetic-based drug strategies.Clin Epigenetics. 2025 Feb 22;17(1):34. doi: 10.1186/s13148-025-01844-w. Clin Epigenetics. 2025. PMID: 39987205 Free PMC article. Review.
-
Navigating the Immune Maze: Pioneering Strategies for Unshackling Cancer Immunotherapy Resistance.Cancers (Basel). 2023 Dec 15;15(24):5857. doi: 10.3390/cancers15245857. Cancers (Basel). 2023. PMID: 38136402 Free PMC article. Review.
-
Management of T-cell malignancies: Bench-to-bedside targeting of epigenetic biology.CA Cancer J Clin. 2025 Jul-Aug;75(4):282-307. doi: 10.3322/caac.70001. Epub 2025 Apr 15. CA Cancer J Clin. 2025. PMID: 40232267 Free PMC article. Review.
-
DNA and RNA Methylation in Rheumatoid Arthritis-A Narrative Review.Epigenomes. 2025 Jan 8;9(1):2. doi: 10.3390/epigenomes9010002. Epigenomes. 2025. PMID: 39846569 Free PMC article. Review.
References
Publication types
LinkOut - more resources
Full Text Sources

