Beyond Soil-Dwelling Actinobacteria: Fantastic Antibiotics and Where to Find Them
- PMID: 35203798
- PMCID: PMC8868522
- DOI: 10.3390/antibiotics11020195
Beyond Soil-Dwelling Actinobacteria: Fantastic Antibiotics and Where to Find Them
Abstract
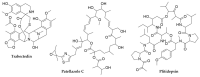
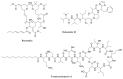
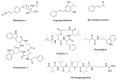
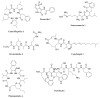
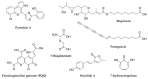
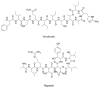
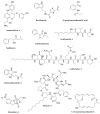
Bacterial secondary metabolites represent an invaluable source of bioactive molecules for the pharmaceutical and agrochemical industries. Although screening campaigns for the discovery of new compounds have traditionally been strongly biased towards the study of soil-dwelling Actinobacteria, the current antibiotic resistance and discovery crisis has brought a considerable amount of attention to the study of previously neglected bacterial sources of secondary metabolites. The development and application of new screening, sequencing, genetic manipulation, cultivation and bioinformatic techniques have revealed several other groups of bacteria as producers of striking chemical novelty. Biosynthetic machineries evolved from independent taxonomic origins and under completely different ecological requirements and selective pressures are responsible for these structural innovations. In this review, we summarize the most important discoveries related to secondary metabolites from alternative bacterial sources, trying to provide the reader with a broad perspective on how technical novelties have facilitated the access to the bacterial metabolic dark matter.
Keywords: antibiotics; bacterial diversity; entomopathogenic bacteria; genome mining; secondary metabolites; symbiosis; unculturable bacteria.
Conflict of interest statement
The authors declare no conflict of interest.
Figures









References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

