Prediction and Modeling of Protein-Protein Interactions Using "Spotted" Peptides with a Template-Based Approach
- PMID: 35204702
- PMCID: PMC8961654
- DOI: 10.3390/biom12020201
Prediction and Modeling of Protein-Protein Interactions Using "Spotted" Peptides with a Template-Based Approach
Abstract
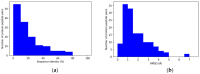
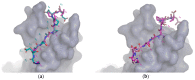
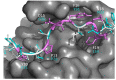
Protein-peptide interactions (PpIs) are a subset of the overall protein-protein interaction (PPI) network in the living cell and are pivotal for the majority of cell processes and functions. High-throughput methods to detect PpIs and PPIs usually require time and costs that are not always affordable. Therefore, reliable in silico predictions represent a valid and effective alternative. In this work, a new algorithm is described, implemented in a freely available tool, i.e., "PepThreader", to carry out PPIs and PpIs prediction and analysis. PepThreader threads multiple fragments derived from a full-length protein sequence (or from a peptide library) onto a second template peptide, in complex with a protein target, "spotting" the potential binding peptides and ranking them according to a sequence-based and structure-based threading score. The threading algorithm first makes use of a scoring function that is based on peptides sequence similarity. Then, a rerank of the initial hits is performed, according to structure-based scoring functions. PepThreader has been benchmarked on a dataset of 292 protein-peptide complexes that were collected from existing databases of experimentally determined protein-peptide interactions. An accuracy of 80%, when considering the top predicted 25 hits, was achieved, which performs in a comparable way with the other state-of-art tools in PPIs and PpIs modeling. Nonetheless, PepThreader is unique in that it is able at the same time to spot a binding peptide within a full-length sequence involved in PPI and model its structure within the receptor. Therefore, PepThreader adds to the already-available tools supporting the experimental PPIs and PpIs identification and characterization.
Keywords: PepThreader; protein–peptide interactions; protein–protein interactions; template-based modeling.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Nilofer C., Sukhwal A., Mohanapriya A., Kangueane P. Open access Volume 13(6) Hypothesis Protein-protein interfaces are vdW dominant with selective H-bonds and (or) electrostatics towards broad functional specificity Open access. Bioinformation. 2017;13:164–173. doi: 10.6026/97320630013164. - DOI - PMC - PubMed
-
- Powell H.R. From then till now: Changing data collection methods in single crystal X-ray crystallography since 1912. Crystallogr. Rev. 2019;25:264–294. doi: 10.1080/0889311X.2019.1615483. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

