The Identification of MATE Antisense Transcripts in Soybean Using Strand-Specific RNA-Seq Datasets
- PMID: 35205273
- PMCID: PMC8871956
- DOI: 10.3390/genes13020228
The Identification of MATE Antisense Transcripts in Soybean Using Strand-Specific RNA-Seq Datasets
Abstract
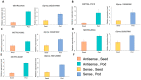
Natural antisense transcripts (NATs) have been generally reported as negative regulators of their sense counterparts. Multidrug and toxic compound extrusion (MATE) proteins mediate the transport of various substrates. Although MATEs have been identified genome-wide in various plant species, their transcript regulators remain unclear. Here, using the publicly available strand-specific RNA-seq datasets of Glycine soja (wild soybean) which have the data from various tissues including developing pods, developing seeds, embryos, cotyledons and hypocotyls, roots, apical buds, stems, and flowers, we identified 35 antisense transcripts of MATEs from 28 gene loci after transcriptome assembly. Spearman correlation coefficients suggested the positive expression correlations of eight MATE antisense and sense transcript pairs. By aligning the identified transcripts with the reference genome of Glycine max (cultivated soybean), the MATE antisense and sense transcript pairs were identified. Using soybean C08 (Glycine max), in developing pods and seeds, the positive correlations between MATE antisense and sense transcript pairs were shown by RT-qPCR. These findings suggest that soybean antisense transcripts are not necessarily negative transcription regulators of their sense counterparts. This study enhances the existing knowledge on the transcription regulation of MATE transporters by uncovering the previously unknown MATE antisense transcripts and their potential synergetic effects on sense transcripts.
Keywords: MATE (multidrug and toxic compound extrusion) transporter; natural antisense transcript (NAT); sense transcript; soybean; strand-specific RNA-seq.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Shoji T., Inai K., Yazaki Y., Sato Y., Takase H., Shitan N., Yazaki K., Goto Y., Toyooka K., Matsuoka K., et al. Multidrug and toxic compound extrusion-type transporters implicated in vacuolar sequestration of nicotine in tobacco roots. Plant Physiol. 2008;149:708–718. doi: 10.1104/pp.108.132811. - DOI - PMC - PubMed
-
- Takanashi K., Shitan N., Yazaki K. The multidrug and toxic compound extrusion (MATE) family in plants. Plant Biotechnol. 2014;31:417–430. doi: 10.5511/plantbiotechnology.14.0904a. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

