Driving Chromatin Organisation through N6-methyladenosine Modification of RNA: What Do We Know and What Lies Ahead?
- PMID: 35205384
- PMCID: PMC8871937
- DOI: 10.3390/genes13020340
Driving Chromatin Organisation through N6-methyladenosine Modification of RNA: What Do We Know and What Lies Ahead?
Abstract
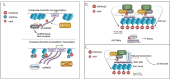
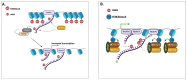
In recent years, there has been an increase in research efforts surrounding RNA modification thanks to key breakthroughs in NGS-based whole transcriptome mapping methods. More than 100 modifications have been reported in RNAs, and some have been mapped at single-nucleotide resolution in the mammalian transcriptome. This has opened new research avenues in fields such as neurobiology, developmental biology, and oncology, among others. To date, we know that the RNA modification machinery finely tunes many diverse mechanisms involved in RNA processing and translation to regulate gene expression. However, it appears obvious to the research community that we have only just begun the process of understanding the several functions of the dynamic web of RNA modification, or the "epitranscriptome". To expand the data generated so far, recently published studies revealed a dual role for N6-methyladenosine (m6A), the most abundant mRNA modification, in driving both chromatin dynamics and transcriptional output. These studies showed that the m6A-modified, chromatin-associated RNAs could act as molecular docks, recruiting histone modification proteins and thus contributing to the regulation of local chromatin structure. Here, we review these latest exciting findings and outline outstanding research questions whose answers will help to elucidate the biological relevance of the m6A modification of chromatin-associated RNAs in mammalian cells.
Keywords: LLPS (Liquid–Liquid Phase Separation); N6-methyladenosine; chromatin; chromatin-associated RNAs; histone modifications; transcription; transposable elements.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

