Engineered Fungus Thermothelomyces thermophilus Producing Plant Storage Proteins
- PMID: 35205873
- PMCID: PMC8877005
- DOI: 10.3390/jof8020119
Engineered Fungus Thermothelomyces thermophilus Producing Plant Storage Proteins
Abstract
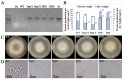
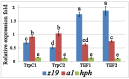
An efficient Agrobacterium-mediated genetic transformation based on the plant binary vector pPZP-RCS2 was carried out for the multiple heterologous protein production in filamentous fungus Thermothelomyces thermophilus F-859 (formerly Myceliophthora thermophila F-859). The engineered fungus Th. thermophilus was able to produce plant storage proteins of Zea mays (α-zein Z19) and Amaranthus hypochondriacus (albumin A1) to enrich fungal biomass by valuable nutritional proteins and improved amino acid content. The mRNA levels of z19 and a1 genes were significantly dependent on their driving promoters: the promoter of tryptophan synthase (PtrpC) was more efficient to express a1, while the promoter of translation elongation factor (Ptef) provided much higher levels of z19 transcript abundance. In general, the total recombinant proteins and amino acid contents were higher in the Ptef-containing clones. This work describes a new strategy to improve mycoprotein nutritive value by overexpression of plant storage proteins.
Keywords: Myceliophthora thermophila; amaranth albumin A1; filamentous fungi; maize alpha-zein; mycoprotein; recombinant protein.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Parisi G., Tulli F., Fortina R., Marino R., Bani P., Dalle Zotte A., De Angelis A., Piccolo G., Pinotti L., Schiavone A., et al. Protein Hunger of the Feed Sector: The Alternatives Offered by the Plant World. Ital. J. Anim. Sci. 2020;19:1204–1225. doi: 10.1080/1828051X.2020.1827993. - DOI
-
- Dhillon G.S. In: Protein Byproducts. Transformation from Environmental Burden into Value-Added Products. Dhillon G.S., editor. Academic Press; Camridge, MA, USA: 2016.
-
- Li M.H., Robinson E.H., Bosworth B.G., Oberle D.F., Lucas P.M. Use of Corn Gluten Feed and Cottonseed Meal to Replace Soybean Meal and Corn in Diets for Pond-Raised Channel Catfish. N. Am. J. Aquac. 2011;73:153–158. doi: 10.1080/15222055.2011.568857. - DOI
-
- Schweiggert-Weisz U., Eisner P., Bader-Mittermaier S., Osen R. Food Proteins from Plants and Fungi. Curr. Opin. Food Sci. 2020;32:156–162. doi: 10.1016/j.cofs.2020.08.003. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

