Reliable and Scalable SARS-CoV-2 qPCR Testing at a High Sample Throughput: Lessons Learned from the Belgian Initiative
- PMID: 35207446
- PMCID: PMC8879918
- DOI: 10.3390/life12020159
Reliable and Scalable SARS-CoV-2 qPCR Testing at a High Sample Throughput: Lessons Learned from the Belgian Initiative
Abstract
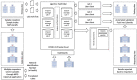
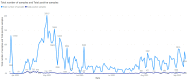
We present our approach to rapidly establishing a standardized, multi-site, nation-wide COVID-19 screening program in Belgium. Under auspices of a federal government Task Force responsible for upscaling the country's testing capacity, we were able to set up a national testing initiative with readily available resources, putting in place a robust, validated, high-throughput, and decentralized qPCR molecular testing platform with embedded proficiency testing. We demonstrate how during an acute scarcity of equipment, kits, reagents, personnel, protective equipment, and sterile plastic supplies, we introduced an approach to rapidly build a reliable, validated, high-volume, high-confidence workflow based on heterogeneous instrumentation and diverse assays, assay components, and protocols. The workflow was set up with continuous quality control monitoring, tied together through a clinical-grade information management platform for automated data analysis, real-time result reporting across different participating sites, qc monitoring, and making result data available to the requesting physician and the patient. In this overview, we address challenges in optimizing high-throughput cross-laboratory workflows with minimal manual intervention through software, instrument and assay validation and standardization, and a process for harmonized result reporting and nation-level infection statistics monitoring across the disparate testing methodologies and workflows, necessitated by a rapid scale-up as a response to the pandemic.
Keywords: SARS-CoV-2; data analysis; high-throughput testing; lab automation; qPCR; qc monitoring.
Conflict of interest statement
Some authors are employees of commercial companies and may own stock, stock options, or other securities in the respective companies. Nathalie Devos, Benjamin Hengchen, Pierre Wattiau, and Sibylle Mehauden are employees of the GSK group of companies. The authors declare no further conflict of interest.
Figures




References
-
- List of Laboratories in Belgium Offering Diagnostic COVID-19 Testing. [(accessed on 21 December 2021)]. Available online: https://covid-19.sciensano.be/sites/default/files/Covid19/COVID-19_Diagn....
-
- Press Release on the COVID-19 Task Force by the Belgium Government. [(accessed on 21 December 2021)]. Available online: https://www.brusselstimes.com/belgium/103427/coronavirus-belgium-looks-t...
-
- WHO COVID-19 Health System Response Monitor. [(accessed on 21 December 2021)]. Available online: https://eurohealthobservatory.who.int/monitors/hsrm/
-
- Tang Y.-W., Schmitz J.E., Persing D.H., Stratton C.W. Laboratory Diagnosis of COVID-19: Current Issues and Challenges. [(accessed on 23 June 2021)];J. Clin. Microbiol. 2020 58:e00512-20. doi: 10.1128/JCM.00512-20. Available online: https://journals.asm.org/doi/abs/10.1128/jcm.00512-20. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

