Is the Alpha Variant of SARS-CoV-2 Associated with a Higher Viral Load than the Historical Strain in Saliva Samples in Patients with Mild to Moderate Symptoms?
- PMID: 35207451
- PMCID: PMC8879902
- DOI: 10.3390/life12020163
Is the Alpha Variant of SARS-CoV-2 Associated with a Higher Viral Load than the Historical Strain in Saliva Samples in Patients with Mild to Moderate Symptoms?
Abstract
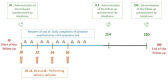
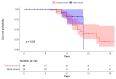
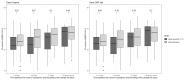
During the COVID-19 pandemic, several generic variants emerged, including the Alpha variant, with increased transmissibility compared to historical strains. We aimed to compare the evolution of the viral load between patients infected with the Alpha variant and those infected with the historical SARS-CoV-2 strains, while taking into account the time interval between the onset of symptoms and samples. We used data collected from patients with an acute respiratory infection (mild to moderate symptoms) and seen in consultation in primary care, included in a prospective longitudinal study, COVID-A. Patients performed four salivary samples during the follow-up. All patients who had at least one of the saliva samples test positive for SARS-CoV-2 were included in the analysis. Overall, 118 patients were included: 89 infected by the historical strain and 29 infected by the Alpha variant. Even though we tended to observe a higher viral load in the Alpha variant group, we found no significant difference in the evolution of the viral load in saliva samples between patients infected with the Alpha variant of the SARS-CoV-2 and those infected by historical strains when controlling for the time interval between the onset of symptoms and sampling.
Keywords: COVID-19; SARS-CoV-2; primary care; variant; viral load.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Gaymard A., Bosetti P., Feri A., Destras G., Enouf V., Andronico A., Burrel S., Behillil S., Sauvage C., Bal A., et al. Early assessment of diffusion and possible expansion of SARS-CoV-2 Lineage 20I/501Y.V1 (B.1.1.7, variant of concern 202012/01) in France, January to March 2021. Eurosurveillance. 2021;26:2100133. doi: 10.2807/1560-7917.ES.2021.26.9.2100133. - DOI - PMC - PubMed
-
- Rambaut A., Loman N., Pybus O., Barclay W., Barrett J., Carabelli A., Connor T., Peacock T., Robertson D., Volz E., et al. Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations. Genom. Epidemiol. 2020 pre-print .
-
- Bal A., Destras G., Gaymard A., Stefic K., Marlet J., Eymieux S., Regue H., Semanas Q., d’Aubarede C., Billaud G., et al. Two-step strategy for the identification of SARS-CoV-2 variant of concern 202012/01 and other variants with spike deletion H69-V70, France, August to December 2020. Eurosurveillance. 2021;26:2100008. doi: 10.2807/1560-7917.ES.2021.26.3.2100008. - DOI - PMC - PubMed
-
- Davies N.G., Abbott S., Barnard R.C., Jarvis C.I., Kucharski A.J., Munday J.D., Pearson C.A.B., Russell T.W., Tully D.C., Washburne A.D., et al. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science. 2021;372:eabg3055. doi: 10.1126/science.abg3055. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

