High Expression of Interferon Pathway Genes CXCL10 and STAT2 Is Associated with Activated T-Cell Signature and Better Outcome of Oral Cancer Patients
- PMID: 35207629
- PMCID: PMC8877377
- DOI: 10.3390/jpm12020140
High Expression of Interferon Pathway Genes CXCL10 and STAT2 Is Associated with Activated T-Cell Signature and Better Outcome of Oral Cancer Patients
Abstract
To improve the survival rate of cancer patients, biomarkers for both early diagnosis and patient stratification for appropriate therapeutics play crucial roles in precision oncology. Investigation of altered gene expression and the relevant molecular pathways in cancer cells are helpful for discovering such biomarkers. In this study, we explore the potential prognostic biomarkers for oral cancer patients through systematically analyzing five oral cancer transcriptomic data sets (TCGA, GSE23558, GSE30784, GSE37991, and GSE138206). Gene Set Enrichment Analysis (GSEA) was individually applied to each data set and the upregulated Hallmark molecular pathways of each data set were intersected to generate 13 common pathways including interferon-α/γ pathways. Among the 5 oral cancer data sets, 43 interferon pathway genes were commonly upregulated and 17 genes exhibited prognostic values in TCGA cohort. After validating in another oral cancer cohort (GSE65858), high expressions of C-X-C motif chemokine ligand 10 (CXCL10) and Signal transducer and activator of transcription 2 (STAT2) were confirmed to be good prognostic biomarkers. GSEA of oral cancers stratified by CXCL10/STAT2 expression showed that activation of T-cell pathways and increased tumor infiltration scores of Type 1 T helper (Th1) and CD8+ T cells were associated with high CXCL10/STAT2 expression. These results suggest that high CXCL10/STAT2 expression can predict a favorable outcome in oral cancer patients.
Keywords: CXCL10; STAT2; T cells; biomarker; interferon; oral cancer; prognosis.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures






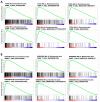
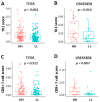
Similar articles
-
Chemokine (C-X-C motif) ligand (CXCL)10 in autoimmune diseases.Autoimmun Rev. 2014 Mar;13(3):272-80. doi: 10.1016/j.autrev.2013.10.010. Epub 2013 Nov 2. Autoimmun Rev. 2014. PMID: 24189283 Review.
-
CXCL10 is a potential biomarker and associated with immune infiltration in human papillary thyroid cancer.Biosci Rep. 2021 Jan 29;41(1):BSR20203459. doi: 10.1042/BSR20203459. Biosci Rep. 2021. PMID: 33345267 Free PMC article.
-
A novel prognostic model for osteosarcoma using circulating CXCL10 and FLT3LG.Cancer. 2017 Jan 1;123(1):144-154. doi: 10.1002/cncr.30272. Epub 2016 Aug 16. Cancer. 2017. PMID: 27529817 Free PMC article.
-
IL-12, but not IFN-alpha, promotes STAT4 activation and Th1 development in murine CD4+ T cells expressing a chimeric murine/human Stat2 gene.J Immunol. 2005 Jan 1;174(1):294-301. doi: 10.4049/jimmunol.174.1.294. J Immunol. 2005. PMID: 15611252
-
A Positive Feedback Amplifier Circuit That Regulates Interferon (IFN)-Stimulated Gene Expression and Controls Type I and Type II IFN Responses.Front Immunol. 2018 May 28;9:1135. doi: 10.3389/fimmu.2018.01135. eCollection 2018. Front Immunol. 2018. PMID: 29892288 Free PMC article. Review.
Cited by
-
STAT2 Controls Colorectal Tumorigenesis and Resistance to Anti-Cancer Drugs.Cancers (Basel). 2023 Nov 15;15(22):5423. doi: 10.3390/cancers15225423. Cancers (Basel). 2023. PMID: 38001683 Free PMC article.
-
Oncogenic STAT Transcription Factors as Targets for Cancer Therapy: Innovative Strategies and Clinical Translation.Cancers (Basel). 2024 Mar 31;16(7):1387. doi: 10.3390/cancers16071387. Cancers (Basel). 2024. PMID: 38611065 Free PMC article. Review.
-
The duality of STAT2 mediated type I interferon signaling in the tumor microenvironment and chemoresistance.Cytokine. 2023 Jan;161:156081. doi: 10.1016/j.cyto.2022.156081. Epub 2022 Oct 31. Cytokine. 2023. PMID: 36327541 Free PMC article. Review.
-
STAT2 act a prognostic biomarker and associated with immune infiltration in kidney renal clear cell carcinoma.Medicine (Baltimore). 2023 Apr 25;102(17):e33662. doi: 10.1097/MD.0000000000033662. Medicine (Baltimore). 2023. PMID: 37115061 Free PMC article.
-
Dual role of CXCL10 in cancer progression: implications for immunotherapy and targeted treatment.Cancer Biol Ther. 2025 Dec;26(1):2538962. doi: 10.1080/15384047.2025.2538962. Epub 2025 Aug 4. Cancer Biol Ther. 2025. PMID: 40760734 Free PMC article. Review.
References
-
- Vermorken J.B., Budach V., Leemans C.R., Machiels J.-P., Nicolai P., O’Sullivan B. Critical Issues in Head and Neck Oncology: Key Concepts from the Seventh THNO Meeting. Springer; Cham, Switzerland: 2021. p. 1. SpringerLink (Online Service)
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

