Expression of Selected Genes and Circulating microRNAs in Patients with Celiac Disease
- PMID: 35208504
- PMCID: PMC8878253
- DOI: 10.3390/medicina58020180
Expression of Selected Genes and Circulating microRNAs in Patients with Celiac Disease
Abstract
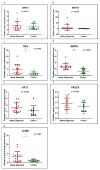
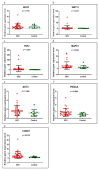
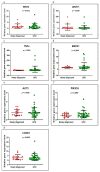
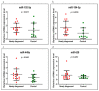
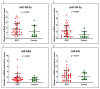
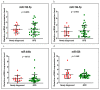
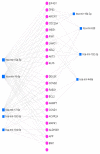
Background and Objectives: Celiac disease (CD) is an immune-mediated enteropathy with characteristic intestinal alterations. CD occurs as a chronic inflammation secondary to gluten sensitivity in genetically susceptible individuals. Until now, the exact cause of the disease has not been established, which is why new studies have appeared that address the involvement of various genes and microRNAs (miRNAs) in the pathogenesis. The aim of the study is to describe the expression of selected genes (Wnt family member 3, WNT3; Wnt family member 11, WNT11; tumor necrosis factor alpha, TNFα; mitogen-activated protein kinase 1, MAPK1; AKT serine/threonine kinase 3, AKT3; phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha, PIK3CA; and cyclin D1, CCND1) and miRNAs (miR-192-5p, miR-194-5p, miR-449a and miR-638) in adult patients with CD. Materials and Methods: In total, 15 patients with CD at diagnosis (newly diagnosed), 33 patients on a gluten-free diet (GFD) for at least 1 year and 10 controls (control) were prospectively included. Blood samples were evaluated by quantitative real-time polymerase chain reaction (qRT-PCR). Results: The results show that TNFα, MAPK1 and CCND1 were significantly overexpressed (p = 0.0249, p = 0.0019 and p = 0.0275, respectively) when comparing the newly diagnosed group to the controls. The other genes studied in CD patients were mostly with high values compared to controls, without reaching statistical significance. Among the miRNAs, the closest to a statistically significant value was miR-194-5p when the newly diagnosed group versus control (p = 0.0510) and GFD group versus control (p = 0.0671) were compared. The DIANA and miRNet databases identified significant functional activity for miR-449a and miR-192-5p and an interconnection of miR-194-5p and miR-449a with CCND1. Conclusions: In conclusion, genes and circulating miRNAs require further studies as they could represent important biomarkers in clinical practice.
Keywords: biomarkers; celiac disease; circulating microRNAs; gene expression; networks.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Expression of MicroRNAs in Adults with Celiac Disease: A Narrative Review.Int J Mol Sci. 2024 Aug 30;25(17):9412. doi: 10.3390/ijms25179412. Int J Mol Sci. 2024. PMID: 39273359 Free PMC article. Review.
-
Circulating microRNAs as novel non-invasive biomarkers of paediatric celiac disease and adherence to gluten-free diet.EBioMedicine. 2022 Feb;76:103851. doi: 10.1016/j.ebiom.2022.103851. Epub 2022 Feb 9. EBioMedicine. 2022. PMID: 35151110 Free PMC article.
-
Altered miRNA expression in duodenal tissue of celiac patients and the impact of a gluten-free diet: a preliminary study.Mol Biol Rep. 2025 Apr 30;52(1):441. doi: 10.1007/s11033-025-10534-y. Mol Biol Rep. 2025. PMID: 40304865 Free PMC article.
-
Circulating miRNAs as Potential Biomarkers for Celiac Disease Development.Front Immunol. 2021 Dec 7;12:734763. doi: 10.3389/fimmu.2021.734763. eCollection 2021. Front Immunol. 2021. PMID: 34950132 Free PMC article.
-
Intestinal and Circulating MicroRNAs in Coeliac Disease.Int J Mol Sci. 2017 Sep 6;18(9):1907. doi: 10.3390/ijms18091907. Int J Mol Sci. 2017. PMID: 28878141 Free PMC article. Review.
Cited by
-
Role and Significance of MicroRNAs in the Relationship Between Obesity and Cancer.Balkan Med J. 2025 May 5;42(3):188-200. doi: 10.4274/balkanmedj.galenos.2025.2025-3-60. Balkan Med J. 2025. PMID: 40326803 Free PMC article. Review.
-
Biomarkers to Monitor Adherence to Gluten-Free Diet by Celiac Disease Patients: Gluten Immunogenic Peptides and Urinary miRNAs.Foods. 2022 May 10;11(10):1380. doi: 10.3390/foods11101380. Foods. 2022. PMID: 35626950 Free PMC article. Review.
-
Expression of MicroRNAs in Adults with Celiac Disease: A Narrative Review.Int J Mol Sci. 2024 Aug 30;25(17):9412. doi: 10.3390/ijms25179412. Int J Mol Sci. 2024. PMID: 39273359 Free PMC article. Review.
-
Circulating microRNAs Suggest Networks Associated with Biological Functions in Aggressive Refractory Type 2 Celiac Disease.Biomedicines. 2022 Jun 14;10(6):1408. doi: 10.3390/biomedicines10061408. Biomedicines. 2022. PMID: 35740429 Free PMC article.
-
Malignancies in Patients with Celiac Disease: Diagnostic Challenges and Molecular Advances.Genes (Basel). 2023 Jan 31;14(2):376. doi: 10.3390/genes14020376. Genes (Basel). 2023. PMID: 36833303 Free PMC article. Review.
References
-
- Al-Toma A., Volta U., Auricchio R., Castillejo G., Sanders D.S., Cellier C., Mulder C.J., Lundin K.E. European Society for the Study of Coeliac Disease (ESsCD) guideline for coeliac disease and other gluten-related disorders. United Eur. Gastroenterol. J. 2019;7:583–613. doi: 10.1177/2050640619844125. - DOI - PMC - PubMed
-
- Domsa E.M., Filip G.A., Olteanu D., Baldea I., Clichici S., Muresan A., David L., Moldovan B., Suciu M., Achim M., et al. Gold nanoparticles phytoreduced with Cornus mas extract mitigate some of gliadin effects on Caco-2 cells. J. Physiol. Pharmacol. 2020;71:201–212. doi: 10.26402/jpp.2020.2.04. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

