A Polyphasic Approach Reveals Novel Genotypes and Updates the Genetic Structure of the Banana Fusarium Wilt Pathogen
- PMID: 35208723
- PMCID: PMC8876670
- DOI: 10.3390/microorganisms10020269
A Polyphasic Approach Reveals Novel Genotypes and Updates the Genetic Structure of the Banana Fusarium Wilt Pathogen
Abstract
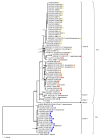
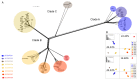
Fusarium oxysporum f. sp. cubense (Foc) is a soil-borne fungus that causes Fusarium wilt, a destructive plant disease that has resulted in devastating economic losses to banana production worldwide. The fungus has a complex evolutionary history and taxonomic repute and consists of three pathogenic races and at least 24 vegetative compatibility groups (VCGs). Surveys conducted in Asia, Africa, the Sultanate of Oman and Mauritius encountered isolates of F. oxysporum pathogenic to banana that were not compatible to any of the known Foc VCGs. Genetic relatedness between the undescribed and known Foc VCGs were determined using a multi-gene phylogeny and diversity array technology (DArT) sequencing. The presence of putative effector genes, the secreted in xylem (SIX) genes, were also determined. Fourteen novel Foc VCGs and 17 single-member VCGs were identified. The multi-gene tree was congruent with the DArT-seq phylogeny and divided the novel VCGs into three clades. Clustering analysis of the DArT-seq data supported the separation of Foc isolates into eight distinct clusters, with the suite of SIX genes mostly conserved within these clusters. Results from this study indicates that Foc is more diverse than hitherto assumed.
Keywords: banana Fusarium wilt; diversity array technology; phylogeny; secreted in xylem genes; vegetative compatibility.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Leslie J.F., Summerell B.A. The Fusarium Laboratory Manual. Blackwell Publishing; Ames, IA, USA: 2006.
-
- Su H.J. Fusarial Wilt of Cavendish Bananas in Taiwan. Plant Dis. 1986;70:814–818. doi: 10.1094/pd-70-814. - DOI
-
- Pegg K., Moore N., Bentley S. Fusarium wilt of banana in Australia: A review. Aust. J. Agric. Res. 1996;47:637–650. doi: 10.1071/AR9960637. - DOI
-
- Ploetz R., Pegg K. Fusarium wilt of banana and Wallace’s line: Was the disease originally restricted to his Indo-Malayan region? Australas. Plant Pathol. 1997;26:239–249. doi: 10.1071/AP97039. - DOI
LinkOut - more resources
Full Text Sources

