BODIPY Conjugates as Functional Compounds for Medical Diagnostics and Treatment
- PMID: 35209191
- PMCID: PMC8877204
- DOI: 10.3390/molecules27041396
BODIPY Conjugates as Functional Compounds for Medical Diagnostics and Treatment
Abstract
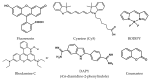
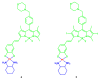
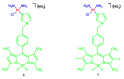
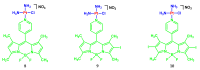
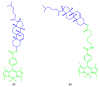
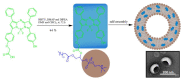
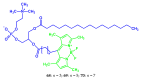
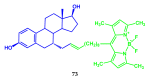
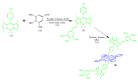
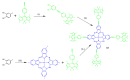
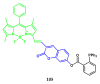
Fluorescent dyes absorbing and emitting in the visible and near-IR regions are promising for the development of fluorescent probes for labeling and bio-visualization of body cells. The ability to absorb and emit in the long-wavelength region increases the efficiency of recording the spectral signals of the probes due to the higher permeability of the skin layers. Compared to other fluorescent dyes, BODIPYs are attractive due to their excellent photophysical properties-narrow absorption and emission, intense fluorescence, simple signal modulation for the practical applications. As part of conjugates with biomolecules, BODIPY could act as a biomarker, but as therapeutic agent, which allows solving several problems at once-labeling or bioimaging and treatment based on the suppression of pathogenic microflora and cancer cells, which provides a huge potential for practical application of BODIPY conjugates in medicine. The review is devoted to the discussion of the recent, promising directions of BODIPY application in the field of conjugation with biomolecules. The first direction is associated with the development of BODIPY conjugates with drugs, including compounds of platinum, paclitaxel, chlorambucil, isoxazole, capsaicin, etc. The second direction is devoted to the labeling of vitamins, hormones, lipids, and other biomolecules to control the processes of their transport, localization in target cells, and metabolism. Within the framework of the third direction, the problem of obtaining functional optically active materials by conjugating BODIPY with other colored and fluorescent particles, in particular, phthalocyanines, is being solved.
Keywords: BODIPY; biomarker; conjugate; drug delivery; fluorescent label; luminophore.
Conflict of interest statement
The authors declare no conflict of interest.
Figures














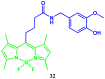
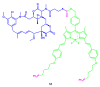
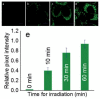

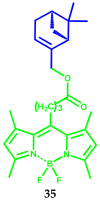
















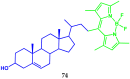
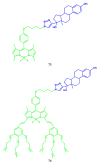
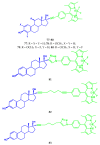
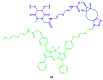










Similar articles
-
Far-red and near infrared BODIPY dyes: synthesis and applications for fluorescent pH probes and bio-imaging.Org Biomol Chem. 2014 Jun 21;12(23):3774-91. doi: 10.1039/c3ob42554a. Org Biomol Chem. 2014. PMID: 24781214 Review.
-
Aromatic [b]-fused BODIPY dyes as promising near-infrared dyes.Org Biomol Chem. 2020 Jun 10;18(22):4135-4156. doi: 10.1039/d0ob00790k. Org Biomol Chem. 2020. PMID: 32441725 Review.
-
Recent Advances of Individual BODIPY and BODIPY-Based Functional Materials in Medical Diagnostics and Treatment.Curr Med Chem. 2017;24(25):2745-2772. doi: 10.2174/0929867324666170601092327. Curr Med Chem. 2017. PMID: 28571557 Review.
-
Fluorescent Labeling of Protein Using Blue-Emitting 8-Amino-BODIPY Derivatives.J Fluoresc. 2017 Nov;27(6):2231-2238. doi: 10.1007/s10895-017-2164-5. Epub 2017 Aug 19. J Fluoresc. 2017. PMID: 28823107
-
Conventional BODIPY Conjugates for Live-Cell Super-Resolution Microscopy and Single-Molecule Tracking.J Vis Exp. 2020 Jun 8;(160):10.3791/60950. doi: 10.3791/60950. J Vis Exp. 2020. PMID: 32568221 Free PMC article.
Cited by
-
Novel carboranyl-BODIPY conjugates: design, synthesis and anti-cancer activity.RSC Adv. 2024 Oct 29;14(47):34643-34660. doi: 10.1039/d4ra07241c. eCollection 2024 Oct 29. RSC Adv. 2024. PMID: 39479484 Free PMC article.
-
Stimuli-Responsive Boron-Based Materials in Drug Delivery.Int J Mol Sci. 2023 Feb 1;24(3):2757. doi: 10.3390/ijms24032757. Int J Mol Sci. 2023. PMID: 36769081 Free PMC article. Review.
-
Investigation of simple BODIPY dyes as G-quadruplex recognizing ligands.RSC Adv. 2025 Feb 17;15(7):5220-5231. doi: 10.1039/d4ra08464k. eCollection 2025 Feb 13. RSC Adv. 2025. PMID: 39963466 Free PMC article.
-
Electropolymerization on ITO-Coated Glass Slides of a Series of π-Extended BODIPY Dyes with Redox-Active Meso-Substituents.Molecules. 2023 Dec 15;28(24):8101. doi: 10.3390/molecules28248101. Molecules. 2023. PMID: 38138589 Free PMC article.
-
Revealing tumor cells and tissues with high selectivity through folic acid-targeted nanofluorescence probes responsive to acidic microenvironments.Front Oncol. 2024 Jun 12;14:1404148. doi: 10.3389/fonc.2024.1404148. eCollection 2024. Front Oncol. 2024. PMID: 38933449 Free PMC article.
References
-
- Deng Z., Wang N., Ai F., Wang Z., Zhu G. Nanomaterial-mediated platinum drug-based combinatorial cancer therapy. View. 2021;2:20200030. doi: 10.1002/VIW.20200030. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

