Circulating microbial content in myeloid malignancy patients is associated with disease subtypes and patient outcomes
- PMID: 35210415
- PMCID: PMC8873459
- DOI: 10.1038/s41467-022-28678-x
Circulating microbial content in myeloid malignancy patients is associated with disease subtypes and patient outcomes
Abstract
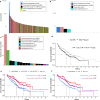
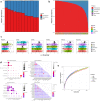
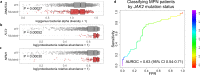
Although recent work has described the microbiome in solid tumors, microbial content in hematological malignancies is not well-characterized. Here we analyze existing deep DNA sequence data from the blood and bone marrow of 1870 patients with myeloid malignancies, along with healthy controls, for bacterial, fungal, and viral content. After strict quality filtering, we find evidence for dysbiosis in disease cases, and distinct microbial signatures among disease subtypes. We also find that microbial content is associated with host gene mutations and with myeloblast cell percentages. In patients with low-risk myelodysplastic syndrome, we provide evidence that Epstein-Barr virus status refines risk stratification into more precise categories than the current standard. Motivated by these observations, we construct machine-learning classifiers that can discriminate among disease subtypes based solely on bacterial content. Our study highlights the association between the circulating microbiome and patient outcome, and its relationship with disease subtype.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Arber DA, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127:2391–2405. - PubMed
-
- Visser O, et al. Incidence, survival and prevalence of myeloid malignancies in Europe. Eur. J. Cancer. 2012;48:3257–3266. - PubMed
-
- Shallis RM, Wang R, Davidoff A, Ma X, Zeidan AM. Epidemiology of acute myeloid leukemia: recent progress and enduring challenges. Blood Rev. 2019;36:70–87. - PubMed
-
- de Martel C, et al. Global burden of cancers attributable to infections in 2008: a review and synthetic analysis. Lancet Oncol. 2012;13:607–615. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

