Identifying Loci Under Positive Selection in Yellow Korean Cattle (Hanwoo)
- PMID: 35210744
- PMCID: PMC8862131
- DOI: 10.1177/1176934319859001
Identifying Loci Under Positive Selection in Yellow Korean Cattle (Hanwoo)
Abstract
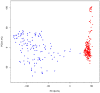
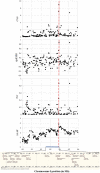
Jeju Black cattle is one of the aboriginal Korean cattle breeds that has been isolated in Jeju island for a long time, while Yellow Hanwoo cattle has been extensively selected for beef production traits for the last several decades. Aiming to investigate broader patterns of selection, we genotyped 352 Yellow Hanwoo and 169 Jeju Black cattle using a customized 150 K bovine chip. Our composite selection signals' analysis to identify selection signatures (cross-population extended haplotype homozygosity [XP-EHH], ΔSAF, and F ST) identified recent and strong signature of selection near many loci with mutations affecting the traits under strong selection as outlier in Yellow Hanwoo, including SCP2 (P = 8.41 × 10-10) that may be involved in the meat quality. We found nine candidate regions with significant clusters of selection signals, and further bioinformatics analyses of the genes located within these regions revealed mainly genes involved in G-protein coupled receptor signaling pathway (GO:0007186) or olfactory transduction (bta04740), which may be due to adaptation to natural environments in Jeju island. Based on the stronger correlation of Ne10/Ne100 ratio between Yellow Hanwoo (0.61) and Jeju Black (0.66) cattle, our results suggest that the difference of chromosomal regions of selection signature between the 2 cattle breeds was due to a consequence of selection processes to adapt to environmental differences between Jeju island and the main inland, Korean peninsula.
Keywords: Jeju black cattle; Yellow Hanwoo; effective population size; positive selection.
© The Author(s) 2019.
Conflict of interest statement
Declaration of conflicting interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Figures


Similar articles
-
Comparisons of Beef Fatty Acid and Amino Acid Characteristics between Jeju Black Cattle, Hanwoo, and Wagyu Breeds.Food Sci Anim Resour. 2019 Jun;39(3):402-409. doi: 10.5851/kosfa.2019.e33. Epub 2019 Jun 30. Food Sci Anim Resour. 2019. PMID: 31304469 Free PMC article.
-
Genetic characteristics of Korean Jeju Black cattle with high density single nucleotide polymorphisms.Anim Biosci. 2021 May;34(5):789-800. doi: 10.5713/ajas.19.0888. Epub 2020 Aug 21. Anim Biosci. 2021. PMID: 32882779 Free PMC article.
-
Genomic Footprints in Selected and Unselected Beef Cattle Breeds in Korea.PLoS One. 2016 Mar 29;11(3):e0151324. doi: 10.1371/journal.pone.0151324. eCollection 2016. PLoS One. 2016. PMID: 27023061 Free PMC article.
-
Towards breed formation by island model divergence in Korean cattle.BMC Evol Biol. 2015 Dec 18;15:284. doi: 10.1186/s12862-015-0563-2. BMC Evol Biol. 2015. PMID: 26677975 Free PMC article.
-
Hanwoo cattle: origin, domestication, breeding strategies and genomic selection.J Anim Sci Technol. 2014 May 15;56:2. doi: 10.1186/2055-0391-56-2. eCollection 2014. J Anim Sci Technol. 2014. PMID: 26290691 Free PMC article. Review.
Cited by
-
Assessing Population Structure and Signatures of Selection in Wanbei Pigs Using Whole Genome Resequencing Data.Animals (Basel). 2022 Dec 20;13(1):13. doi: 10.3390/ani13010013. Animals (Basel). 2022. PMID: 36611624 Free PMC article.
References
LinkOut - more resources
Full Text Sources

