Head on Comparison of Self- and Nano-assemblies of Gamma Peptide Nucleic Acid Amphiphiles
- PMID: 35210986
- PMCID: PMC8863176
- DOI: 10.1002/adfm.202109552
Head on Comparison of Self- and Nano-assemblies of Gamma Peptide Nucleic Acid Amphiphiles
Abstract
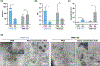
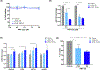
Peptide nucleic acids (PNAs) are nucleic acid analogs with superior hybridization properties and enzymatic stability than deoxyribonucleic acid (DNA). In addition to gene targeting applications, PNAs have garnered significant attention as bio-polymer due to the Watson-Crick -based molecular recognition and flexibility of synthesis. Here, we engineered PNA amphiphiles using chemically modified gamma PNA (8 mer in length) containing hydrophilic diethylene glycol units at the gamma position and covalently conjugated lauric acid (C12) as a hydrophobic moiety. Gamma PNA (γPNA) amphiphiles self-assemble into spherical vesicles. Further, we formulate nano-assemblies using the amphiphilic γPNA as a polymer via ethanol injection-based protocols. We perform comprehensive head-on comparison of the physicochemical and cellular uptake properties of PNA derived self- and nano-assemblies. Small-angle neutron scattering (SANS) and small-angle X-ray scattering (SAXS) analysis reveal ellipsoidal morphology of γPNA nano-assemblies that results in superior cellular delivery compate to the spherical self-assembly. Next, we compare the functional activities of γPNA self-and nano-assemblies in lymphoma cells via multiple endpoints, including gene expression, cell viability, and apoptosis-based assays. Overall, we establish that γPNA amphiphile is a functionally active bio-polymer to formulate nano-assemblies for a wide range of biomedical applications.
Keywords: PNA amphiphiles; gamma peptide nucleic acids (γPNAs); micro-RNA-155; nano-assemblies; self-assembly.
Conflict of interest statement
Conflict of Interest Authors declare no conflict of interest.
Figures




Similar articles
-
Head-to-head comparison of in vitro and in vivo efficacy of pHLIP-conjugated anti-seed gamma peptide nucleic acids.Cell Rep Phys Sci. 2023 Oct 18;4(10):101584. doi: 10.1016/j.xcrp.2023.101584. Epub 2023 Sep 15. Cell Rep Phys Sci. 2023. PMID: 38144419 Free PMC article.
-
Chimeric γPNA-Invader probes: using intercalator-functionalized oligonucleotides to enhance the DNA-targeting properties of γPNA.Org Biomol Chem. 2020 Feb 21;18(7):1359-1368. doi: 10.1039/c9ob02726b. Epub 2020 Jan 27. Org Biomol Chem. 2020. PMID: 31984413
-
DNA-π Amphiphiles: A Unique Building Block for the Crafting of DNA-Decorated Unilamellar Nanostructures.Acc Chem Res. 2020 Nov 17;53(11):2668-2679. doi: 10.1021/acs.accounts.0c00492. Epub 2020 Oct 14. Acc Chem Res. 2020. PMID: 33052654
-
Molecular self-assembly using peptide nucleic acids.Biopolymers. 2017 Jan;108(1):10.1002/bip.22930. doi: 10.1002/bip.22930. Biopolymers. 2017. PMID: 27486924 Free PMC article. Review.
-
Engineered Aptamer-Organic Amphiphile Self-Assemblies for Biomedical Applications: Progress and Challenges.Small. 2022 Jan;18(4):e2104341. doi: 10.1002/smll.202104341. Epub 2021 Oct 7. Small. 2022. PMID: 34622570 Review.
Cited by
-
Recent Cutting-Edge Technologies for the Delivery of Peptide Nucleic Acid.Chemistry. 2025 Jun 17;31(34):e202500469. doi: 10.1002/chem.202500469. Epub 2025 May 21. Chemistry. 2025. PMID: 40351137 Free PMC article. Review.
-
Functionalized Peptide-Based Nanoparticles for Targeted Cancer Nanotherapeutics: A State-of-the-Art Review.ACS Omega. 2022 Oct 5;7(41):36092-36107. doi: 10.1021/acsomega.2c03974. eCollection 2022 Oct 18. ACS Omega. 2022. PMID: 36278104 Free PMC article. Review.
-
Unlocking the potential of chemically modified peptide nucleic acids for RNA-based therapeutics.RNA. 2023 Apr;29(4):434-445. doi: 10.1261/rna.079498.122. Epub 2023 Jan 18. RNA. 2023. PMID: 36653113 Free PMC article. Review.
-
Advances in Nucleic Acid Research: Exploring the Potential of Oligonucleotides for Therapeutic Applications and Biological Studies.Int J Mol Sci. 2023 Dec 21;25(1):146. doi: 10.3390/ijms25010146. Int J Mol Sci. 2023. PMID: 38203317 Free PMC article. Review.
-
Next-generation poly-L-histidine formulations for miRNA mimic delivery.Mol Ther Methods Clin Dev. 2023 Apr 1;29:271-283. doi: 10.1016/j.omtm.2023.03.015. eCollection 2023 Jun 8. Mol Ther Methods Clin Dev. 2023. PMID: 37123088 Free PMC article.
References
-
- Nielsen PE, Egholm M, Berg RH, Buchardt O, Science 1991, 254, 1497. - PubMed
-
- Wittung P, Nielsen PE, Buchardt O, Egholm M, Norden B, Nature 1994, 368, 561. - PubMed
-
- Demidov VV, Potaman VN, Frank-Kamenetskii MD, Egholm M, Buchard O, Sonnichsen SH, Nielsen PE, Biochem Pharmacol 1994, 48, 1310. - PubMed
-
- Egholm M, Buchardt O, Christensen L, Behrens C, Freier SM, Driver DA, Berg RH, Kim SK, Norden B, Nielsen PE, Nature 1993, 365, 566. - PubMed
-
- Bahal R, Ali McNeer N, Quijano E, Liu Y, Sulkowski P, Turchick A, Lu YC, Bhunia DC, Manna A, Greiner DL, Brehm MA, Cheng CJ, Lopez-Giraldez F, Ricciardi A, Beloor J, Krause DS, Kumar P, Gallagher PG, Braddock DT, Mark Saltzman W, Ly DH, Glazer PM, Nat Commun 2016, 7, 13304; - PMC - PubMed
- Bahal R, Quijano E, McNeer NA, Liu Y, Bhunia DC, Lopez-Giraldez F, Fields RJ, Saltzman WM, Ly DH, Glazer PM, Curr Gene Ther 2014, 14, 331; - PMC - PubMed
- Ricciardi AS, Bahal R, Farrelly JS, Quijano E, Bianchi AH, Luks VL, Putman R, Lopez-Giraldez F, Coskun S, Song E, Liu Y, Hsieh WC, Ly DH, Stitelman DH, Glazer PM, Saltzman WM, Nat Commun 2018, 9, 2481. - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
