Mapping Floral Genetic Architecture in Prunus mume, an Ornamental Woody Plant
- PMID: 35211141
- PMCID: PMC8860970
- DOI: 10.3389/fpls.2022.828579
Mapping Floral Genetic Architecture in Prunus mume, an Ornamental Woody Plant
Abstract
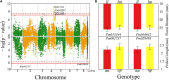
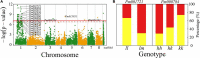
Floral traits are both evolutionarily and economically relevant for ornamental plants. However, their underlying genetic architecture, especially in woody ornamental plants, is still poorly understood. We perform mapping experiments aimed at identifying specific quantitative trait loci (QTLs) that control the size, shape, architecture, color, and timing of flowers in mei (Prunus mume). We find that the narrow region of chromosome 1 (5-15 Mb) contains a number of floral QTLs. Most QTLs detected from this mapping study are annotated to candidate genes that regulate various biological functions toward the floral formation. We identify strong pleiotropic control on different aspects of flower morphology (including shape, petal number, pistil number, petal color, and calyx color) and flower timing, but find different genetic systems that mediate whether a flower produces pistils and how many pistils a flower produces. We find that many floral QTLs display pleiotropic effects on shoot length growth but shoot radial growth, implicating a possible association of floral display with light capture. We conduct a transcriptomic study to characterize the genomic signature of floral QTLs expressed in mei. Our mapping results about the genetic control of floral features make it promising to select superior varieties for mei carrying flowers of ornamental value.
Keywords: Prunus mume (mei); QTL; floral trait; genetic mapping; pleiotropy; transcriptomic analysis.
Copyright © 2022 Li, Sang, Wen, Meng, Cheng, Zhang and Sun.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures






Similar articles
-
The genetic architecture of floral traits in the woody plant Prunus mume.Nat Commun. 2018 Apr 27;9(1):1702. doi: 10.1038/s41467-018-04093-z. Nat Commun. 2018. PMID: 29703940 Free PMC article.
-
Genetic control of juvenile growth and botanical architecture in an ornamental woody plant, Prunus mume Sieb. et Zucc. as revealed by a high-density linkage map.BMC Genet. 2014;15 Suppl 1(Suppl 1):S1. doi: 10.1186/1471-2156-15-S1-S1. Epub 2014 Jun 20. BMC Genet. 2014. PMID: 25078672 Free PMC article.
-
A Computational Model for Inferring QTL Control Networks Underlying Developmental Covariation.Front Plant Sci. 2019 Dec 18;10:1557. doi: 10.3389/fpls.2019.01557. eCollection 2019. Front Plant Sci. 2019. PMID: 31921232 Free PMC article.
-
Towards the Improvement of Ornamental Attributes in Chrysanthemum: Recent Progress in Biotechnological Advances.Int J Mol Sci. 2022 Oct 14;23(20):12284. doi: 10.3390/ijms232012284. Int J Mol Sci. 2022. PMID: 36293140 Free PMC article. Review.
-
Biotechnological Advancements for Improving Floral Attributes in Ornamental Plants.Front Plant Sci. 2017 Apr 20;8:530. doi: 10.3389/fpls.2017.00530. eCollection 2017. Front Plant Sci. 2017. PMID: 28473834 Free PMC article. Review.
Cited by
-
A High-Resolution Linkage Map Construction and QTL Analysis for Morphological Traits in Anthurium (Anthurium andraeanum Linden).Plants (Basel). 2023 Dec 17;12(24):4185. doi: 10.3390/plants12244185. Plants (Basel). 2023. PMID: 38140512 Free PMC article.
-
Exploring the wild almond, Prunus arabica (Olivier), as a genetic source for almond breeding.Tree Genet Genomes. 2024;20(5):37. doi: 10.1007/s11295-024-01668-4. Epub 2024 Sep 24. Tree Genet Genomes. 2024. PMID: 39398478 Free PMC article.
-
Genomic region and origin for selected traits during differentiation of small-fruit cultivars in Japanese apricot (Prunus mume).Mol Genet Genomics. 2023 Nov;298(6):1365-1375. doi: 10.1007/s00438-023-02062-w. Epub 2023 Aug 26. Mol Genet Genomics. 2023. PMID: 37632570
-
An integrated QTL and RNA-seq analysis revealed new petal morphology loci in Brassica napus L.Biotechnol Biofuels Bioprod. 2024 Jul 18;17(1):105. doi: 10.1186/s13068-024-02551-z. Biotechnol Biofuels Bioprod. 2024. PMID: 39026359 Free PMC article.
-
Mutations overlying the miR172 target site of TOE-type genes are prime candidate variants for the double-flower trait in mei.Sci Rep. 2024 Mar 27;14(1):7300. doi: 10.1038/s41598-024-57589-8. Sci Rep. 2024. PMID: 38538684 Free PMC article.
References
-
- Bernardello G., Anderson G. J., Stuessy T. F., Crawford D. J. (2001). A survey of floral traits, breeding systems, floral visitors, and pollination systems of the angiosperms of the Juan Fernández Islands (Chile). Bot. Rev. 67 255–308. 10.1007/BF02858097 - DOI
LinkOut - more resources
Full Text Sources
Miscellaneous

