Isolation and characterization mesenchymal stem cells from red panda (Ailurus fulgens styani) endometrium
- PMID: 35211318
- PMCID: PMC8862722
- DOI: 10.1093/conphys/coac004
Isolation and characterization mesenchymal stem cells from red panda (Ailurus fulgens styani) endometrium
Abstract
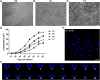
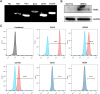
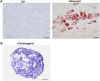
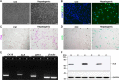
Endometrial mesenchymal stem cells (eMSCs) are undifferentiated endometrial cells with self-renewal, multidirectional differentiation and high proliferation potential. Nowadays, eMSCs have been found in a few species, but it has never been reported in endangered wild animals, especially the red panda. In this study, we successfully isolated and characterized the eMSCs derived from red panda. Red panda eMSCs were fibroblast-like, had a strong proliferative potential and a stable chromosome number. Pluripotency genes including Klf4, Sox2 and Thy1 were highly expressed in eMSCs. Besides, cultured eMSCs were positive for MSC markers CD44, CD49f and CD105 and negative for endothelial cell marker CD31 and haematopoietic cell marker CD34. Moreover, no reference RNA-seq was used to analyse the eMSCs transcriptional expression profile and key pathways. Compared with skin fibroblast cell group, 9104 differentially expressed genes (DEGs) were identified, among which are 5034 genes upregulated, 4070 genes downregulated and the top 20 enrichment pathways of DEGs in Gene Ontology (GO) and the Kyoto Encyclopedia of Genes Genomes (KEGG) mainly associated with G-protein coupled receptor signalling pathway, carbohydrate derivative binding, nucleoside binding, ribosome biogenesis, cell cycle, DNA replication, Ras signalling pathway and purine metabolism. Among the DEGs, some representative genes about promoting MSCs differentiation and proliferation were upregulated and promoting fibroblasts proliferation were downregulated in eMSCs group. Red panda eMSCs also had multiple differentiation ability and could differentiate into adipocytes, chondrocytes and hepatocytes. In conclusion, we, for the first time, isolated and characterized the red panda eMSCs with ability of multiplication and multilineage differentiation in vitro. The new multipotential stem cell could be beneficial not only for the germ plasm resources conservation of red panda, but also for basic or pre-clinical studies in the future.
Keywords: Cell culture; cell differentiation; endometrium; mesenchymal stem cell; red panda.
© The Author(s) 2022. Published by Oxford University Press and the Society for Experimental Biology.
Figures

References
-
- An JH, Li FP, He P, Chen JS, Cai ZG, Liu SR, Yue CJ, Liu YL, Hou R (2020) Characteristics of mesenchymal stem cells isolated from the bone marrow of red pandas. Zoology (Jena) 140: 125775. - PubMed
-
- Bian Y, Du Y, Wang R, Chen N, Du X, Wang Y, Yuan H (2019) A comparative study of HAMSCs/HBMSCs transwell and mixed coculture systems. IUBMB Life 71: 1048–1055. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

