Human Stool Preservation Impacts Taxonomic Profiles in 16S Metagenomics Studies
- PMID: 35211421
- PMCID: PMC8860989
- DOI: 10.3389/fcimb.2022.722886
Human Stool Preservation Impacts Taxonomic Profiles in 16S Metagenomics Studies
Abstract
Microbiotas play critical roles in human health, yet in most cases scientists lack standardized and reproducible methods from collection and preservation of samples, as well as the choice of omic analysis, up to the data processing. To date, stool sample preservation remains a source of technological bias in metagenomic sequencing, despite newly developed storage solutions. Here, we conducted a comparative study of 10 storage methods for human stool over a 14-day period of storage at fluctuating temperatures. We first compared the performance of each stabilizer with observed bacterial composition variation within the same specimen. Then, we identified the nature of the observed variations to determine which bacterial populations were more impacted by the stabilizer. We found that DNA stabilizers display various stabilizing efficacies and affect the recovered bacterial profiles thus highlighting that some solutions are more performant in preserving the true gut microbial community. Furthermore, our results showed that the bias associated with the stabilizers can be linked to the phenotypical traits of the bacterial populations present in the studied samples. Although newly developed storage solutions have improved our capacity to stabilize stool microbial content over time, they are nevertheless not devoid of biases hence requiring the implantation of standard operating procedures. Acknowledging the biases and limitations of the implemented method is key to better interpret and support true associated microbiome patterns that will then lead us towards personalized medicine, in which the microbiota profile could constitute a reliable tool for clinical practice.
Keywords: 16S metagenomics; human gut; microbiota; preservation; stabilizing solution.; standardization; stool.
Copyright © 2022 Plauzolles, Toumi, Bonnet, Pénaranda, Bidaut, Chiche, Allardet-Servent, Retornaz, Goutorbe and Halfon.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


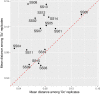
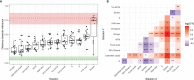
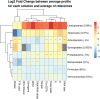
References
-
- Brenner D. J., Staley J. T., Krieg N. R. (2005). “Classification of Procaryotic Organisms and the Concept of Bacterial Speciation,” in Bergey’s Manual® of Systematic Bacteriology: Volume Two: The Proteobacteria, Part A Introductory Essays. Eds. Brenner D. J., Krieg N. R., Staley J. T., Garrity G. M. (Boston, MA: Springer US; ), 27–32. doi: 10.1007/0-387-28021-9_4 - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

