Optimization of Mechanical Tissue Dissociation Using an Integrated Microfluidic Device for Improved Generation of Single Cells Following Digestion
- PMID: 35211466
- PMCID: PMC8861371
- DOI: 10.3389/fbioe.2022.841046
Optimization of Mechanical Tissue Dissociation Using an Integrated Microfluidic Device for Improved Generation of Single Cells Following Digestion
Abstract
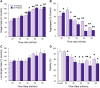
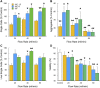
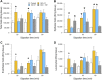
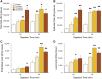
The dissociation of tissue and cell aggregates into single cells is of high interest for single cell analysis studies, primary cultures, tissue engineering, and regenerative medicine. However, current methods are slow, poorly controlled, variable, and can introduce artifacts. We previously developed a microfluidic device that contains two separate dissociation modules, a branching channel array and nylon mesh filters, which was used as a polishing step after tissue processing with a microfluidic digestion device. Here, we employed the integrated disaggregation and filtration (IDF) device as a standalone method with both cell aggregates and traditionally digested tissue to perform a well-controlled and detailed study into the effect of mechanical forces on dissociation, including modulation of flow rate, device pass number, and even the mechanism. Using a strongly cohesive cell aggregate model, we found that single cell recovery was highest using flow rates exceeding 40 ml/min and multiple passes through the filter module, either with or without the channel module. For minced and digested kidney tissue, recovery of diverse cell types was maximal using multiple passes through the channel module and only a single pass through the filter module. Notably, we found that epithelial cell recovery from the optimized IDF device alone exceeded our previous efforts, and this result was maintained after reducing digestion time to 20 min. However, endothelial cells and leukocytes still required extended digestion time for maximal recover. These findings highlight the significance of parameter optimization to achieve the highest cell yield and viability based on tissue sample size, extracellular matrix content, and strength of cell-cell interactions.
Keywords: dissociation; microfluidics; single cell analysis; tissue engineering; tissue processing.
Copyright © 2022 Aliaghaei and Haun.
Conflict of interest statement
JH is a co-founder of Kino Discovery, which is in the process of licensing intellectual property for the tissue processing devices. The remaining author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Microfluidic filter device with nylon mesh membranes efficiently dissociates cell aggregates and digested tissue into single cells.Lab Chip. 2018 Sep 11;18(18):2776-2786. doi: 10.1039/c8lc00507a. Lab Chip. 2018. PMID: 30090895 Free PMC article.
-
Microfluidic device for mechanical dissociation of cancer cell aggregates into single cells.Lab Chip. 2015 Jan 7;15(1):339-350. doi: 10.1039/c4lc01126k. Lab Chip. 2015. PMID: 25377468 Free PMC article.
-
Microfluidic Device Technologies for Digestion, Disaggregation, and Filtration of Tissue Samples for Single Cell Applications.Methods Mol Biol. 2022;2394:81-92. doi: 10.1007/978-1-0716-1811-0_6. Methods Mol Biol. 2022. PMID: 35094323
-
Upflow anaerobic sludge blanket reactor--a review.Indian J Environ Health. 2001 Apr;43(2):1-82. Indian J Environ Health. 2001. PMID: 12397675 Review.
-
Microfluidic strategies for design and assembly of microfibers and nanofibers with tissue engineering and regenerative medicine applications.Adv Healthc Mater. 2015 Jan 7;4(1):11-28. doi: 10.1002/adhm.201400144. Epub 2014 May 23. Adv Healthc Mater. 2015. PMID: 24853649 Review.
Cited by
-
Investigation of the Effect of High Shear Stress on Mesenchymal Stem Cells Using a Rotational Rheometer in a Small-Angle Cone-Plate Configuration.Bioengineering (Basel). 2024 Oct 11;11(10):1011. doi: 10.3390/bioengineering11101011. Bioengineering (Basel). 2024. PMID: 39451387 Free PMC article.
-
Drosophila CASK regulates brain size and neuronal morphogenesis, providing a genetic model of postnatal microcephaly suitable for drug discovery.Neural Dev. 2023 Oct 7;18(1):6. doi: 10.1186/s13064-023-00174-y. Neural Dev. 2023. PMID: 37805506 Free PMC article.
-
Integrated Microfluidics for Single-Cell Separation and On-Chip Analysis: Novel Applications and Recent Advances.Small Sci. 2024 Feb 2;4(4):2300206. doi: 10.1002/smsc.202300206. eCollection 2024 Apr. Small Sci. 2024. PMID: 40212991 Free PMC article.
-
Improving Transcriptome Fidelity Following Synovial Tissue Disaggregation.Front Med (Lausanne). 2022 Aug 10;9:919748. doi: 10.3389/fmed.2022.919748. eCollection 2022. Front Med (Lausanne). 2022. PMID: 36035425 Free PMC article.
-
Aging murine peritoneal macrophages undergo female-specific remodeling driven by hormone-dependent and independent mechanisms.bioRxiv [Preprint]. 2025 Jun 27:2025.06.11.659200. doi: 10.1101/2025.06.11.659200. bioRxiv. 2025. PMID: 40666991 Free PMC article. Preprint.
References
-
- Ahmed O., Abdellah H., Elsayed M., Abdelgawad M., Mousa N. A., El-Badri N. (2014). “Tissue Dissociation Miniaturized Platform for Uterine Stem Cell Isolation and Culture,” in Cairo International Biomedical Engineering Conference, Giza, Egypt, 11-13 Dec. 2014 (IEEE; ), 178–180. 10.1109/cibec.2014.7020950 - DOI

