Single-cell genome-wide concurrent haplotyping and copy-number profiling through genotyping-by-sequencing
- PMID: 35212381
- PMCID: PMC9226495
- DOI: 10.1093/nar/gkac134
Single-cell genome-wide concurrent haplotyping and copy-number profiling through genotyping-by-sequencing
Erratum in
-
Correction to 'Single-cell genome-wide concurrent haplotyping and copy-number profiling through genotyping-by-sequencing'.Nucleic Acids Res. 2022 Sep 9;50(16):9597. doi: 10.1093/nar/gkac729. Nucleic Acids Res. 2022. PMID: 35979958 Free PMC article. No abstract available.
Abstract
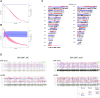
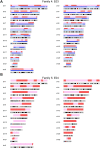
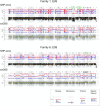
Single-cell whole-genome haplotyping allows simultaneous detection of haplotypes associated with monogenic diseases, chromosome copy-numbering and subsequently, has revealed mosaicism in embryos and embryonic stem cells. Methods, such as karyomapping and haplarithmisis, were deployed as a generic and genome-wide approach for preimplantation genetic testing (PGT) and are replacing traditional PGT methods. While current methods primarily rely on single-nucleotide polymorphism (SNP) array, we envision sequencing-based methods to become more accessible and cost-efficient. Here, we developed a novel sequencing-based methodology to haplotype and copy-number profile single cells. Following DNA amplification, genomic size and complexity is reduced through restriction enzyme digestion and DNA is genotyped through sequencing. This single-cell genotyping-by-sequencing (scGBS) is the input for haplarithmisis, an algorithm we previously developed for SNP array-based single-cell haplotyping. We established technical parameters and developed an analysis pipeline enabling accurate concurrent haplotyping and copy-number profiling of single cells. We demonstrate its value in human blastomere and trophectoderm samples as application for PGT for monogenic disorders. Furthermore, we demonstrate the method to work in other species through analyzing blastomeres of bovine embryos. Our scGBS method opens up the path for single-cell haplotyping of any species with diploid genomes and could make its way into the clinic as a PGT application.
© The Author(s) 2022. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



References
-
- Chen X., Love J.C., Navin N.E., Pachter L, Stubbington M.J.T., Svensson V., Sweedler J.V., Teichmann S.A.. Single-cell analysis at the threshold. Nat. Biotechnol. 2016; 34:1111–1118. - PubMed
-
- Vermeesch J.R., Voet T., Devriendt K.. Prenatal and pre-implantation genetic diagnosis. Nat. Rev. Genet. 2016; 17:643–656. - PubMed
-
- Natesan S.A., Bladon A.J., Coskun S., Qubbaj W., Prates R., Munne S., Coonen E., Dreesen J.C.F.M., Stevens S.J.C., Paulussen A.D.C.et al. .. Genome-wide karyomapping accurately identifies the inheritance of single-gene defects in human preimplantation embryos in vitro. Genet. Med. 2014; 16:838–845. - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

