Mapping the developing human cardiac endothelium at single-cell resolution identifies MECOM as a regulator of arteriovenous gene expression
- PMID: 35212715
- PMCID: PMC9648824
- DOI: 10.1093/cvr/cvac023
Mapping the developing human cardiac endothelium at single-cell resolution identifies MECOM as a regulator of arteriovenous gene expression
Abstract
Aims: Coronary vasculature formation is a critical event during cardiac development, essential for heart function throughout perinatal and adult life. However, current understanding of coronary vascular development has largely been derived from transgenic mouse models. The aim of this study was to characterize the transcriptome of the human foetal cardiac endothelium using single-cell RNA sequencing (scRNA-seq) to provide critical new insights into the cellular heterogeneity and transcriptional dynamics that underpin endothelial specification within the vasculature of the developing heart.
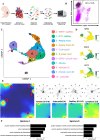
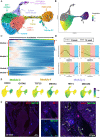
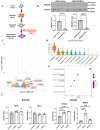
Methods and results: We acquired scRNA-seq data of over 10 000 foetal cardiac endothelial cells (ECs), revealing divergent EC subtypes including endocardial, capillary, venous, arterial, and lymphatic populations. Gene regulatory network analyses predicted roles for SMAD1 and MECOM in determining the identity of capillary and arterial populations, respectively. Trajectory inference analysis suggested an endocardial contribution to the coronary vasculature and subsequent arterialization of capillary endothelium accompanied by increasing MECOM expression. Comparative analysis of equivalent data from murine cardiac development demonstrated that transcriptional signatures defining endothelial subpopulations are largely conserved between human and mouse. Comprehensive characterization of the transcriptional response to MECOM knockdown in human embryonic stem cell-derived EC (hESC-EC) demonstrated an increase in the expression of non-arterial markers, including those enriched in venous EC.
Conclusions: scRNA-seq of the human foetal cardiac endothelium identified distinct EC populations. A predicted endocardial contribution to the developing coronary vasculature was identified, as well as subsequent arterial specification of capillary EC. Loss of MECOM in hESC-EC increased expression of non-arterial markers, suggesting a role in maintaining arterial EC identity.
Keywords: Coronary vasculature formation; Endothelial heterogeneity; Human cardiac development; MECOM; Single-cell RNA sequencing; Vascular regeneration.
© The Author(s) 2022. Published by Oxford University Press on behalf of the European Society of Cardiology.
Conflict of interest statement
Conflict of interest: none declared.
Figures


Comment in
-
A new resource for human coronary vessel development.Cardiovasc Res. 2022 Nov 10;118(14):2875-2876. doi: 10.1093/cvr/cvac094. Cardiovasc Res. 2022. PMID: 35726909 Free PMC article. No abstract available.
References
-
- Wu B, Zhang Z, Lui W, Chen X, Wang Y, Chamberlain AA, Moreno-Rodriguez RA, Markwald RR, O'Rourke BP, Sharp DJ, Zheng D, Lenz J, Baldwin HS, Chang C-P, Zhou B. Endocardial cells form the coronary arteries by angiogenesis through myocardial-endocardial VEGF signaling. Cell 2012;151:1083–1096. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

