Protein Dynamics to Define and Refine Disordered Protein Ensembles
- PMID: 35213160
- PMCID: PMC10122607
- DOI: 10.1021/acs.jpcb.1c10925
Protein Dynamics to Define and Refine Disordered Protein Ensembles
Abstract
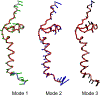
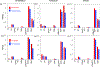
Intrinsically disordered proteins and unfolded proteins have fluctuating conformational ensembles that are fundamental to their biological function and impact protein folding, stability, and misfolding. Despite the importance of protein dynamics and conformational sampling, time-dependent data types are not fully exploited when defining and refining disordered protein ensembles. Here we introduce a computational framework using an elastic network model and normal-mode displacements to generate a dynamic disordered ensemble consistent with NMR-derived dynamics parameters, including transverse R2 relaxation rates and Lipari-Szabo order parameters (S2 values). We illustrate our approach using the unfolded state of the drkN SH3 domain to show that the dynamical ensembles give better agreement than a static ensemble for a wide range of experimental validation data including NMR chemical shifts, J-couplings, nuclear Overhauser effects, paramagnetic relaxation enhancements, residual dipolar couplings, hydrodynamic radii, single-molecule fluorescence Förster resonance energy transfer, and small-angle X-ray scattering.
Figures
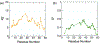

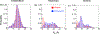
References
-
- Wright PE; Dyson HJ Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm. Journal of molecular biology 1999, 293 (2), 321–331. - PubMed
-
- Uversky VN Intrinsically disordered proteins from A to Z. The international journal of biochemistry & cell biology 2011, 43 (8), 1090–1103. - PubMed
-
- Tompa P.Intrinsically disordered proteins: a 10-year recap. Trends in biochemical sciences 2012, 37 (12), 509–516. - PubMed
-
- Uversky VN Unusual biophysics of intrinsically disordered proteins. Biochimica et Biophysica Acta (BBA)-Proteins and Proteomics 2013, 1834 (5), 932–951. - PubMed

