Therapeutic antibody activation of the glucocorticoid-induced TNF receptor by a clustering mechanism
- PMID: 35213218
- PMCID: PMC8880771
- DOI: 10.1126/sciadv.abm4552
Therapeutic antibody activation of the glucocorticoid-induced TNF receptor by a clustering mechanism
Abstract
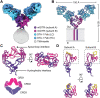
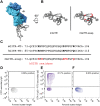
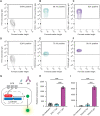
GITR is a TNF receptor, and its activation promotes immune responses and drives antitumor activity. The receptor is activated by the GITR ligand (GITRL), which is believed to cluster receptors into a high-order array. Immunotherapeutic agonist antibodies also activate the receptor, but their mechanisms are not well characterized. We solved the structure of full-length mouse GITR bound to Fabs from the antibody DTA-1. The receptor is a dimer, and each subunit binds one Fab in an orientation suggesting that the antibody clusters receptors. Binding experiments with purified proteins show that DTA-1 IgG and GITRL both drive extensive clustering of GITR. Functional data reveal that DTA-1 and the anti-human GITR antibody TRX518 activate GITR in their IgG forms but not as Fabs. Thus, the divalent character of the IgG agonists confers an ability to mimic GITRL and cluster and activate GITR. These findings will inform the clinical development of this class of antibodies for immuno-oncology.
Figures



Similar articles
-
HERA-GITRL activates T cells and promotes anti-tumor efficacy independent of FcγR-binding functionality.J Immunother Cancer. 2019 Jul 19;7(1):191. doi: 10.1186/s40425-019-0671-4. J Immunother Cancer. 2019. PMID: 31324216 Free PMC article.
-
GITR ligand fusion protein agonist enhances the tumor antigen-specific CD8 T-cell response and leads to long-lasting memory.J Immunother Cancer. 2017 Jun 20;5:47. doi: 10.1186/s40425-017-0247-0. eCollection 2017. J Immunother Cancer. 2017. PMID: 28649380 Free PMC article.
-
Structures of mouse and human GITR-GITRL complexes reveal unique TNF superfamily interactions.Nat Commun. 2021 Mar 2;12(1):1378. doi: 10.1038/s41467-021-21563-z. Nat Commun. 2021. PMID: 33654081 Free PMC article.
-
Pharmacological modulation of GITRL/GITR system: therapeutic perspectives.Br J Pharmacol. 2012 Apr;165(7):2089-99. doi: 10.1111/j.1476-5381.2011.01753.x. Br J Pharmacol. 2012. PMID: 22029729 Free PMC article. Review.
-
GITR-GITRL system, a novel player in shock and inflammation.ScientificWorldJournal. 2007 May 1;7:533-66. doi: 10.1100/tsw.2007.106. ScientificWorldJournal. 2007. PMID: 17525820 Free PMC article. Review.
Cited by
-
Combinations of anti-GITR antibody and CD28 superagonist ameliorated dextran sodium sulfate-induced mouse colitis.Clin Exp Immunol. 2022 Jun 23;208(3):340-350. doi: 10.1093/cei/uxac039. Clin Exp Immunol. 2022. PMID: 35511600 Free PMC article.
-
Phase IB Study of GITR Agonist Antibody TRX518 Singly and in Combination with Gemcitabine, Pembrolizumab, or Nivolumab in Patients with Advanced Solid Tumors.Clin Cancer Res. 2022 Sep 15;28(18):3990-4002. doi: 10.1158/1078-0432.CCR-22-0339. Clin Cancer Res. 2022. PMID: 35499569 Free PMC article.
-
GITR and TIGIT immunotherapy provokes divergent multicellular responses in the tumor microenvironment of gastrointestinal cancers.Genome Med. 2023 Nov 26;15(1):100. doi: 10.1186/s13073-023-01259-3. Genome Med. 2023. PMID: 38008725 Free PMC article.
-
Impact of Glucocorticoids on Cardiovascular System-The Yin Yang Effect.J Pers Med. 2022 Nov 3;12(11):1829. doi: 10.3390/jpm12111829. J Pers Med. 2022. PMID: 36579545 Free PMC article. Review.
References
-
- Vanamee É. S., Faustman D. L., Structural principles of tumor necrosis factor superfamily signaling. Sci. Signal. 11, eaao4910 (2018). - PubMed
-
- Dostert C., Grusdat M., Letellier E., Brenner D., The TNF family of ligands and receptors: Communication modules in the immune system and beyond. Physiol. Rev. 99, 115–160 (2019). - PubMed
-
- Aggarwal B. B., Signalling pathways of the TNF superfamily: A double-edged sword. Nat. Rev. Immunol. 3, 745–756 (2003). - PubMed
-
- Kim H. J., Kim H. Y., Kim B. K., Kim S., Chung D. H., Engagement of glucocorticoid-induced TNF receptor costimulates NKT cell activation in vitro and in vivo. J. Immunol. 176, 3507–3515 (2006). - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

