Metabolic profiling of prostate cancer in skeletal microenvironments identifies G6PD as a key mediator of growth and survival
- PMID: 35213227
- PMCID: PMC8880772
- DOI: 10.1126/sciadv.abf9096
Metabolic profiling of prostate cancer in skeletal microenvironments identifies G6PD as a key mediator of growth and survival
Abstract
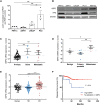
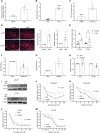
The spread of cancer to bone is invariably fatal, with complex cross-talk between tumor cells and the bone microenvironment responsible for driving disease progression. By combining in silico analysis of patient datasets with metabolomic profiling of prostate cancer cells cultured with bone cells, we demonstrate the changing energy requirements of prostate cancer cells in the bone microenvironment, identifying the pentose phosphate pathway (PPP) as elevated in prostate cancer bone metastasis, with increased expression of the PPP rate-limiting enzyme glucose-6-phosphate dehydrogenase (G6PD) associated with a reduction in progression-free survival. Genetic and pharmacologic manipulation demonstrates that G6PD inhibition reduces prostate cancer growth and migration, associated with changes in cellular redox state and increased chemosensitivity. Genetic blockade of G6PD in vivo results in reduction of tumor growth within bone. In summary, we demonstrate the metabolic plasticity of prostate cancer cells in the bone microenvironment, identifying the PPP and G6PD as metabolic targets for the treatment of prostate cancer bone metastasis.
Figures




References
-
- National Cancer Institute Surveillance, Epidemiology, and End Results Program: Cancer Stat Facts: Prostate Cancer [Internet]. [cited 23 Nov 2020]; https://seer.cancer.gov/statfacts/html/prost.html.
-
- Tangen C. M., Hussain M. H. A., Higano C. S., Eisenberger M. A., Small E. J., Wilding G., Donnelly B. J., Schelhammer P. F., Crawford E. D., Vogelzang N. J., Powell I. J., Thompson I. M. Jr., Improved overall survival trends of men with newly diagnosed M1 prostate cancer: A SWOG phase III trial experience (S8494, S8894 and S9346). J. Urol. 188, 1164–1169 (2012). - PMC - PubMed
-
- Nuhn P., De Bono J. S., Fizazi K., Freedland S. J., Grilli M., Kantoff P. W., Sonpavde G., Sternberg C. N., Yegnasubramanian S., Antonarakis E. S., Update on systemic prostate cancer therapies: Management of metastatic castration-resistant prostate cancer in the era of precision oncology. Eur. Urol. 75, 88–99 (2019). - PubMed
-
- Marin-Aguilera M., Codony-Servat J., Kalko S. G., Fernandez P. L., Bermudo R., Buxo E., Ribal M. J., Gascón P., Mellado B., Identification of docetaxel resistance genes in castration-resistant prostate cancer. Mol. Cancer Ther. 11, 329–339 (2012). - PubMed
-
- Bubendorf L., Schöpfer A., Wagner U., Sauter G., Moch H., Willi N., Gasser T. C., Mihatsch M. J., Metastatic patterns of prostate cancer: An autopsy study of 1,589 patients. Hum. Pathol. 31, 578–583 (2000). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

