HEATR3 variants impair nuclear import of uL18 (RPL5) and drive Diamond-Blackfan anemia
- PMID: 35213692
- PMCID: PMC9136880
- DOI: 10.1182/blood.2021011846
HEATR3 variants impair nuclear import of uL18 (RPL5) and drive Diamond-Blackfan anemia
Abstract
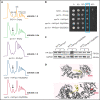
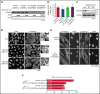
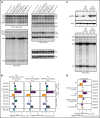
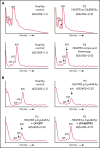
The congenital bone marrow failure syndrome Diamond-Blackfan anemia (DBA) is typically associated with variants in ribosomal protein (RP) genes impairing erythroid cell development. Here we report multiple individuals with biallelic HEATR3 variants exhibiting bone marrow failure, short stature, facial and acromelic dysmorphic features, and intellectual disability. These variants destabilize a protein whose yeast homolog is known to synchronize the nuclear import of RPs uL5 (RPL11) and uL18 (RPL5), which are both critical for producing ribosomal subunits and for stabilizing the p53 tumor suppressor when ribosome biogenesis is compromised. Expression of HEATR3 variants or repression of HEATR3 expression in primary cells, cell lines of various origins, and yeast models impairs growth, differentiation, pre-ribosomal RNA processing, and ribosomal subunit formation reminiscent of DBA models of large subunit RP gene variants. Consistent with a role of HEATR3 in RP import, HEATR3-depleted cells or patient-derived fibroblasts display reduced nuclear accumulation of uL18. Hematopoietic progenitor cells expressing HEATR3 variants or small-hairpin RNAs knocking down HEATR3 synthesis reveal abnormal acceleration of erythrocyte maturation coupled to severe proliferation defects that are independent of p53 activation. Our study uncovers a new pathophysiological mechanism leading to DBA driven by biallelic HEATR3 variants and the destabilization of a nuclear import protein important for ribosome biogenesis.
© 2022 by The American Society of Hematology.
Figures



Comment in
-
Turning up the HEAT(R3) in Diamond-Blackfan anemia.Blood. 2022 May 26;139(21):3101-3102. doi: 10.1182/blood.2022015881. Blood. 2022. PMID: 35616991 No abstract available.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

