Establishment of Sandwich ELISA for Quality Control in Rotavirus Vaccine Production
- PMID: 35214701
- PMCID: PMC8876306
- DOI: 10.3390/vaccines10020243
Establishment of Sandwich ELISA for Quality Control in Rotavirus Vaccine Production
Abstract
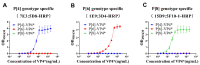
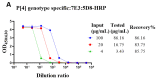
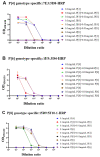
Non-replicating rotavirus vaccines are alternative strategies that may improve the protective efficacy of rotavirus vaccines in low- and middle-income countries. The truncated spike protein VP4 (aa26-476, VP4*)was a candidate antigen for the development of recombinant rotavirus vaccines, with higher immunogenicity and protective efficacy compared to VP8* and VP5* alone. This article describes the development of three genotype-specific sandwich ELISAs for P[4], P[6], and P[8]-VP4*, which are important for quality control in rotavirus vaccine production. Our results showed that the detection systems had good specificity for the different genotype VP4* and were not influenced by the E. coli host proteins. Moreover, the detection systems play an important role in determining whether the target protein was contaminated by VP4* proteins of other genotypes. They can also detect the adsorption rate of the adjuvant to the P[4], P[6], P[8]-VP4* protein during the process development. The three detection systems will play an important role in the quality control and process development of VP4* based rotavirus vaccines and facilitate the development of recombinant rotavirus vaccines.
Keywords: P[4], P[6], P[8]-VP4*; detection; enzyme-linked immunosorbent assay; vaccine.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Troeger C., Forouzanfar M., Rao P.C., Khalil I., Brown A., Reiner R.C., Fullman N., Thompson R.L., Abajobir A., Ahmed M., et al. Estimates of global, regional, and national morbidity, mortality, and aetiologies of diarrhoeal diseases: A systematic analysis for the Global Burden of Disease Study 2015. Lancet Infect. Dis. 2017;17:909–948. doi: 10.1016/S1473-3099(17)30276-1. - DOI - PMC - PubMed
-
- Cunliffe N.A., Witte D., Ngwira B.M., Todd S., Bostock N.J., Turner A.M., Chimpeni P., Victor J.C., Steele A.D., Bouckenooghe A., et al. Efficacy of human rotavirus vaccine against severe gastroenteritis in Malawian children in the first two years of life: A randomized, double-blind, placebo controlled trial. Vaccine. 2012;30((Suppl. 1)):A36–A43. doi: 10.1016/j.vaccine.2011.09.120. - DOI - PMC - PubMed
-
- Zaman K., Anh D.D., Victor J.C., Shin S., Yunus M., Dallas M.J., Podder G., Thiem V.D., Mai L.T.P., Luby S.P., et al. Efficacy of pentavalent rotavirus vaccine against severe rotavirus gastroenteritis in infants in developing countries in Asia: A randomised, double-blind, placebo-controlled trial. Lancet. 2010;376:615–623. doi: 10.1016/S0140-6736(10)60755-6. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

