Immunodominant Linear B-Cell Epitopes of SARS-CoV-2 Spike, Identified by Sera from K18-hACE2 Mice Infected with the WT or Variant Viruses
- PMID: 35214711
- PMCID: PMC8875268
- DOI: 10.3390/vaccines10020251
Immunodominant Linear B-Cell Epitopes of SARS-CoV-2 Spike, Identified by Sera from K18-hACE2 Mice Infected with the WT or Variant Viruses
Abstract
SARS-CoV-2 surface spike protein mediates the viral entry into the host cell and represents the primary immunological target of COVID-19 vaccines as well as post-exposure immunotherapy. Establishment of the highly immunogenic B-cell epitope profile of SARS-CoV-2 proteins in general, and that of the spike protein in particular, may contribute to the development of sensitive diagnostic tools and identification of vaccine` candidate targets. In the current study, the anti-viral antibody response in transgenic K18-hACE-2 mice was examined by implementing an immunodominant epitope mapping approach of the SARS-CoV-2 spike. Serum samples for probing an epitope array covering the entire spike protein were collected from mice following infection with the original SARS-CoV-2 strain as well as the B.1.1.7 Alpha and B.1.351 Beta genetic variants of concern. The analysis resulted in distinction of six linear epitopes common to the humoral response against all virus variants inspected at a frequency of more than 20% of the serum samples. Finally, the universality of the response was probed by cross-protective in vitro experiments using plaque-reducing neutralization tests. The data presented here has important implications for prediction of the efficacy of immune countermeasures against emerging SARS-CoV-2 variants.
Keywords: COVID-19; K18-hACE2; SARS-CoV-2; epitope mapping; linear epitopes.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
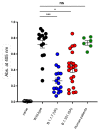
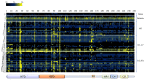

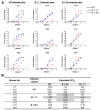
Similar articles
-
A Glycosylated RBD Protein Induces Enhanced Neutralizing Antibodies against Omicron and Other Variants with Improved Protection against SARS-CoV-2 Infection.J Virol. 2022 Sep 14;96(17):e0011822. doi: 10.1128/jvi.00118-22. Epub 2022 Aug 16. J Virol. 2022. PMID: 35972290 Free PMC article.
-
Host immune responses associated with SARS-CoV-2 Omicron infection result in protection or pathology during reinfection depending on mouse genetic background.Res Sq [Preprint]. 2023 Nov 29:rs.3.rs-3637405. doi: 10.21203/rs.3.rs-3637405/v1. Res Sq. 2023. Update in: Nat Commun. 2024 Nov 23;15(1):10178. doi: 10.1038/s41467-024-54334-7. PMID: 38077015 Free PMC article. Updated. Preprint.
-
Comprehensive characterization of the antibody responses to SARS-CoV-2 Spike protein finds additional vaccine-induced epitopes beyond those for mild infection.Elife. 2022 Jan 24;11:e73490. doi: 10.7554/eLife.73490. Elife. 2022. PMID: 35072628 Free PMC article.
-
Identification of B-Cell Epitopes for Eliciting Neutralizing Antibodies against the SARS-CoV-2 Spike Protein through Bioinformatics and Monoclonal Antibody Targeting.Int J Mol Sci. 2022 Apr 14;23(8):4341. doi: 10.3390/ijms23084341. Int J Mol Sci. 2022. PMID: 35457159 Free PMC article. Review.
-
An overview of methods for the structural and functional mapping of epitopes recognized by anti-SARS-CoV-2 antibodies.RSC Chem Biol. 2021 Sep 29;2(6):1580-1589. doi: 10.1039/d1cb00169h. eCollection 2021 Dec 2. RSC Chem Biol. 2021. PMID: 34977572 Free PMC article. Review.
Cited by
-
Dynamics of spike-specific neutralizing antibodies across five-year emerging SARS-CoV-2 variants of concern reveal conserved epitopes that protect against severe COVID-19.Front Immunol. 2025 Feb 18;16:1503954. doi: 10.3389/fimmu.2025.1503954. eCollection 2025. Front Immunol. 2025. PMID: 40040708 Free PMC article.
-
Previous infection with seasonal coronaviruses does not protect male Syrian hamsters from challenge with SARS-CoV-2.Nat Commun. 2023 Sep 26;14(1):5990. doi: 10.1038/s41467-023-41761-1. Nat Commun. 2023. PMID: 37752151 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

