A Systematic Review of T Cell Epitopes Defined from the Proteome of Hepatitis B Virus
- PMID: 35214714
- PMCID: PMC8878595
- DOI: 10.3390/vaccines10020257
A Systematic Review of T Cell Epitopes Defined from the Proteome of Hepatitis B Virus
Abstract
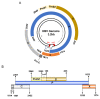
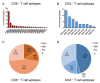
Hepatitis B virus (HBV) infection remains a worldwide health problem and no eradicative therapy is currently available. Host T cell immune responses have crucial influences on the outcome of HBV infection, however the development of therapeutic vaccines, T cell therapies and the clinical evaluation of HBV-specific T cell responses are hampered markedly by the lack of validated T cell epitopes. This review presented a map of T cell epitopes functionally validated from HBV antigens during the past 33 years; the human leukocyte antigen (HLA) supertypes to present these epitopes, and the methods to screen and identify T cell epitopes. To the best of our knowledge, a total of 205 CD8+ T cell epitopes and 79 CD4+ T cell epitopes have been defined from HBV antigens by cellular functional experiments thus far, but most are restricted to several common HLA supertypes, such as HLA-A0201, A2402, B0702, DR04, and DR12 molecules. Therefore, the currently defined T cell epitope repertoire cannot cover the major populations with HLA diversity in an indicated geographic region. More researches are needed to dissect a more comprehensive map of T cell epitopes, which covers overall HBV proteome and global patients.
Keywords: HLA restriction; T cell epitope; hepatitis B virus.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- World Health Organization . Global Hepatitis Report 2017. World Health Organization; Geneva, Switzerland: 2017.
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

