Diversification of Chemical Structures of Methoxylated Flavonoids and Genes Encoding Flavonoid- O-Methyltransferases
- PMID: 35214897
- PMCID: PMC8876552
- DOI: 10.3390/plants11040564
Diversification of Chemical Structures of Methoxylated Flavonoids and Genes Encoding Flavonoid- O-Methyltransferases
Abstract
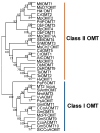
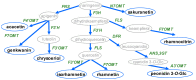
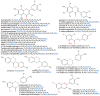
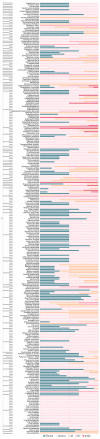
The O-methylation of specialized metabolites in plants is a unique decoration that provides structural and functional diversity of the metabolites with changes in chemical properties and intracellular localizations. The O-methylation of flavonoids, which is a class of plant specialized metabolites, promotes their antimicrobial activities and liposolubility. Flavonoid O-methyltransferases (FOMTs), which are responsible for the O-methylation process of the flavonoid aglycone, generally accept a broad range of substrates across flavones, flavonols and lignin precursors, with different substrate preferences. Therefore, the characterization of FOMTs with the physiology roles of methoxylated flavonoids is useful for crop improvement and metabolic engineering. In this review, we summarized the chemodiversity and physiology roles of methoxylated flavonoids, which were already reported, and we performed a cross-species comparison to illustrate an overview of diversification and conserved catalytic sites of the flavonoid O-methyltransferases.
Keywords: O-methyltransferase; methoxylated flavonoids; plant specialized metabolism.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Molecular cloning and characterization of a flavonoid-O-methyltransferase with broad substrate specificity and regioselectivity from Citrus depressa.BMC Plant Biol. 2016 Aug 22;16(1):180. doi: 10.1186/s12870-016-0870-9. BMC Plant Biol. 2016. PMID: 27549218 Free PMC article.
-
Production of methoxylated flavonoids in yeast using ring A hydroxylases and flavonoid O-methyltransferases from sweet basil.Appl Microbiol Biotechnol. 2018 Jul;102(13):5585-5598. doi: 10.1007/s00253-018-9043-0. Epub 2018 Apr 28. Appl Microbiol Biotechnol. 2018. PMID: 29705956
-
Flavonoid methylation: a novel 4'-O-methyltransferase from Catharanthus roseus, and evidence that partially methylated flavanones are substrates of four different flavonoid dioxygenases.Phytochemistry. 2004 Apr;65(8):1085-94. doi: 10.1016/j.phytochem.2004.02.010. Phytochemistry. 2004. PMID: 15110688
-
Multifunctional 5-hydroxyconiferaldehyde O-methyltransferases (CAldOMTs) in plant metabolism.J Exp Bot. 2024 Mar 14;75(6):1671-1695. doi: 10.1093/jxb/erae011. J Exp Bot. 2024. PMID: 38198655 Review.
-
Diversity and regioselectivity of O-methyltransferases catalyzing the formation of O-methylated flavonoids.Crit Rev Biotechnol. 2024 Sep;44(6):1203-1225. doi: 10.1080/07388551.2023.2280755. Epub 2023 Nov 30. Crit Rev Biotechnol. 2024. PMID: 38035668 Review.
Cited by
-
Stachydrine, a Bioactive Equilibrist for Synephrine, Identified from Four Citrus Chinese Herbs.Molecules. 2023 Apr 29;28(9):3813. doi: 10.3390/molecules28093813. Molecules. 2023. PMID: 37175222 Free PMC article.
-
Metabolome integrated with transcriptome, and genome analysis revealed higher accumulations of phytoalexins enhance resistance against Magnaporthe oryzae in new Zhefang rice variety diantun 506.BMC Plant Biol. 2025 Jul 2;25(1):836. doi: 10.1186/s12870-025-06856-5. BMC Plant Biol. 2025. PMID: 40604494 Free PMC article.
-
Ipecac alkaloid biosynthesis in two evolutionarily distant plants.Nat Chem Biol. 2025 Jun 3. doi: 10.1038/s41589-025-01926-z. Online ahead of print. Nat Chem Biol. 2025. PMID: 40461725
-
Synthesis and Characterization of New Pyrano[2,3-c]pyrazole Derivatives as 3-Hydroxyflavone Analogues.Molecules. 2023 Sep 13;28(18):6599. doi: 10.3390/molecules28186599. Molecules. 2023. PMID: 37764375 Free PMC article.
-
Monofloral Corn Poppy Bee-Collected Pollen-A Detailed Insight into Its Phytochemical Composition and Antioxidant Properties.Antioxidants (Basel). 2023 Jul 14;12(7):1424. doi: 10.3390/antiox12071424. Antioxidants (Basel). 2023. PMID: 37507962 Free PMC article.
References
-
- Ibrahim R., De Luca V., Khouri H., Latchinian L., Brisson L., Charest P. Enzymology and compartmentation of polymethylated flavonol glucosides in Chrysosplenium americanum. Phytochemistry. 1987;26:1237–1245. doi: 10.1016/S0031-9422(00)81789-6. - DOI
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

