Are Methionine Sulfoxide-Containing Proteins Related to Seed Longevity? A Case Study of Arabidopsisthaliana Dry Mature Seeds Using Cyanogen Bromide Attack and Two-Dimensional-Diagonal Electrophoresis
- PMID: 35214905
- PMCID: PMC8875303
- DOI: 10.3390/plants11040569
Are Methionine Sulfoxide-Containing Proteins Related to Seed Longevity? A Case Study of Arabidopsisthaliana Dry Mature Seeds Using Cyanogen Bromide Attack and Two-Dimensional-Diagonal Electrophoresis
Abstract
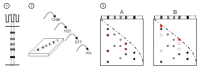
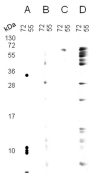
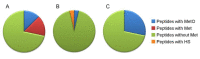
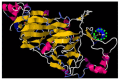
In recent years, several reports pointed out the role of protein oxidation in seed longevity, notably regarding the oxidation of methionine (Met) residues to methionine sulfoxide (MetO) in proteins. To further consider this question, we present a handy proteomic method based on the use of two-dimensional diagonal electrophoresis (2Dd) and cyanogen bromide (CNBr) cleavage, which we refer to as 2Dd-CNBr. CNBr treatment of proteins causes the non-enzymatic hydrolysis of peptide bonds on the carboxyl side of reduced Met residues. However, Met oxidation causes a lack of cleavage, thus modifying the electrophoretic mobility of CNBr-induced peptides. This approach was first validated using bovine serum albumin as a model protein, which confirmed the possibility of distinguishing between oxidized and non-oxidized forms of Met-containing peptides in gels. Then, the 2Dd-CNBr method was applied to the Arabidopsis thaliana seed protein extract in a control (non-oxidized) condition and in an oxidized one (as obtained following hypochlorous acid treatment). Twenty-four oxidized Met residues in 19 proteins identified by mass spectrometry were found to be surface exposed in these proteins. In the three-dimensional environment of the oxidized Met, we detected amino acid residues that could be converted by oxidation (carbonylation) or by phosphorylation, suggesting a possible interplay between Met oxidation and the other protein modifications. The identification of the proteins oxidatively modified in Met residues revealed the finding that MetO-containing proteins are related to seed longevity. Based on these results, we suggest that the method presently described also has the potential for wider applications.
Keywords: mass spectrometry; methionine sulfoxide; oxidative stress; post-translational modifications; protein modification; redox proteomics; seed viability; two-dimensional diagonal electrophoresis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Foyer C.H., Noctor G. Oxidant and Antioxidant Signalling in Plants: A Re-Evaluation of the Concept of Oxidative Stress in a Physiological Context. Plant Cell Environ. 2005;28:1056–1071. doi: 10.1111/j.1365-3040.2005.01327.x. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

