A Lead-Based Fragment Library Screening of the Glycosyltransferase WaaG from Escherichia coli
- PMID: 35215321
- PMCID: PMC8877264
- DOI: 10.3390/ph15020209
A Lead-Based Fragment Library Screening of the Glycosyltransferase WaaG from Escherichia coli
Abstract
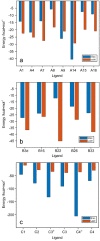
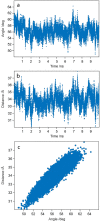
Glucosyl transferase I (WaaG) in E. coli catalyzes the transfer of an α-d-glucosyl group to the inner core of the lipopolysaccharide (LPS) and plays an important role in the biogenesis of the outer membrane. If its activity could be inhibited, the integrity of the outer membrane would be compromised and the bacterium would be susceptible to antibiotics that are normally prevented from entering the cell. Herein, three libraries of molecules (A, B and C) were docked in the binding pocket of WaaG, utilizing the docking binding affinity as a filter to select fragment-based compounds for further investigations. From the results of the docking procedure, a selection of compounds was investigated by molecular dynamics (MD) simulations to obtain binding free energy (BFE) and KD values for ligands as an evaluation for the binding to WaaG. Derivatives of 1,3-thiazoles (A7 and A4) from library A and 1,3,4-thiadiazole (B33) from library B displayed a promising profile of BFE, with KD < mM, viz., 0.11, 0.62 and 0.04 mM, respectively. Further root-mean-square-deviation (RMSD), electrostatic/van der Waals contribution to the binding and H-bond interactions displayed a favorable profile for ligands A4 and B33. Mannose and/or heptose-containing disaccharides C1-C4, representing sub-structures of the inner core of the LPS, were also investigated by MD simulations, and compound C42- showed a calculated KD = 0.4 µM. In the presence of UDP-Glc2-, the best-docked pose of disaccharide C42- is proximate to the glucose-binding site of WaaG. A study of the variation in angle and distance was performed on the different portions of WaaG (N-, the C- domains and the hinge region). The Spearman correlation coefficient between the two variables was close to unity, where both variables increase in the same way, suggesting a conformational rearrangement of the protein during the MD simulation, revealing molecular motions of the enzyme that may be part of the catalytic cycle. Selected compounds were also analyzed by Saturation Transfer Difference (STD) NMR experiments. STD effects were notable for the 1,3-thiazole derivatives A4, A8 and A15 with the apo form of the protein as well as in the presence of UDP for A4.
Keywords: NMR spectroscopy; binding free energy; molecular docking; molecular dynamics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures













References
-
- World Health Organization Prioritization of Pathogens to Guide Discovery, Research and Development of New Antibiotics for Drug Resistant Bacterial Infections, Including Tuberculosis. 2017. [(accessed on 14 January 2022)]. Available online: https://apps.who.int/iris/handle/10665/311820.
-
- Levin S. The crisis in antibiotic resistance. Infect. Dis. Clin. Pract. 1993;2:53. doi: 10.1097/00019048-199301000-00013. - DOI
-
- World Health Organization Outbreaks of E. coli O104:H4 Infection. 2011. [(accessed on 14 January 2022)]. Available online: https://www.euro.who.int/en/countries/germany/outbreaks-of-e.-coli-o104h....
-
- World Health Organization Antimicrobial Resistance. 2020. [(accessed on 14 January 2022)]. Available online: https://www.who.int/health-topics/antimicrobial-resistance.
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

