Translocation, Rejection and Trapping of Polyampholytes
- PMID: 35215709
- PMCID: PMC8877523
- DOI: 10.3390/polym14040797
Translocation, Rejection and Trapping of Polyampholytes
Abstract
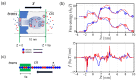
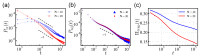
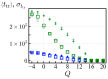
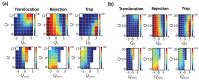
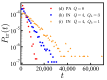
Polyampholytes (PA) are a special class of polymers comprising both positive and negative monomers along their sequence. Most proteins have positive and negative residues and are PAs. Proteins have a well-defined sequence while synthetic PAs have a random charge sequence. We investigated the translocation behavior of random polyampholyte chains through a pore under the action of an electric field by means of Monte Carlo simulations. The simulations incorporated a realistic translocation potential profile along an extended asymmetric pore and translocation was studied for both directions of engagement. The study was conducted from the perspective of statistics for disordered systems. The translocation behavior (translocation vs. rejection) was recorded for all 220 sequences comprised of N = 20 charged monomers. The results were compared with those for 107 random sequences of N = 40 to better demonstrate asymptotic laws. At early times, rejection was mainly controlled by the charge sequence of the head part, but late translocation/rejection was governed by the escape from a trapped state over an antagonistic barrier built up along the sequence. The probability distribution of translocation times from all successful attempts revealed a power-law tail. At finite times, there was a population of trapped sequences that relaxed very slowly (logarithmically) with time. If a subensemble of sequences with prescribed net charge was considered the power-law decay was steeper for a more favorable net charge. Our findings were rationalized by theoretical arguments developed for long chains. We also provided operational criteria for the translocation behavior of a sequence, explaining the selection by the translocation process. From the perspective of protein translocation, our findings can help rationalize the behavior of intrinsically disordered proteins (IDPs), which can be modeled as polyampholytes. Most IDP sequences have a strong net charge favoring translocation. Even for sequences with those large net charges, the translocation times remained very dispersed and the translocation was highly sequence-selective.
Keywords: Monte Carlo simulation; drift-diffusion; polyampholytes; probability distribution function; translocation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Grants and funding
LinkOut - more resources
Full Text Sources

