Identifying Inhibitors of -1 Programmed Ribosomal Frameshifting in a Broad Spectrum of Coronaviruses
- PMID: 35215770
- PMCID: PMC8876150
- DOI: 10.3390/v14020177
Identifying Inhibitors of -1 Programmed Ribosomal Frameshifting in a Broad Spectrum of Coronaviruses
Abstract
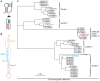
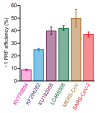
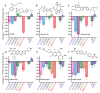
Recurrent outbreaks of novel zoonotic coronavirus (CoV) diseases in recent years have highlighted the importance of developing therapeutics with broad-spectrum activity against CoVs. Because all CoVs use -1 programmed ribosomal frameshifting (-1 PRF) to control expression of key viral proteins, the frameshift signal in viral mRNA that stimulates -1 PRF provides a promising potential target for such therapeutics. To test the viability of this strategy, we explored whether small-molecule inhibitors of -1 PRF in SARS-CoV-2 also inhibited -1 PRF in a range of bat CoVs-the most likely source of future zoonoses. Six inhibitors identified in new and previous screens against SARS-CoV-2 were evaluated against the frameshift signals from a panel of representative bat CoVs as well as MERS-CoV. Some drugs had strong activity against subsets of these CoV-derived frameshift signals, while having limited to no effect on -1 PRF caused by frameshift signals from other viruses used as negative controls. Notably, the serine protease inhibitor nafamostat suppressed -1 PRF significantly for multiple CoV-derived frameshift signals. These results suggest it is possible to find small-molecule ligands that inhibit -1 PRF specifically in a broad spectrum of CoVs, establishing frameshift signals as a viable target for developing pan-coronaviral therapeutics.
Keywords: SARS-CoV-2; coronavirus; programmed ribosomal frameshifting; therapeutics; translation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Comment in
-
Nafamostat-Mediated Inhibition of SARS-CoV-2 Ribosomal Frameshifting Is Insufficient to Impair Viral Replication in Vero Cells. Comment on Munshi et al. Identifying Inhibitors of -1 Programmed Ribosomal Frameshifting in a Broad Spectrum of Coronaviruses. Viruses 2022, 14, 177.Viruses. 2022 Jul 13;14(7):1526. doi: 10.3390/v14071526. Viruses. 2022. PMID: 35891506 Free PMC article.
References
-
- Brierley I., Gilbert R.J., Pennell S. Recoding: Expansion of Decoding Rules Enriches Gene Expression. Springer; New York, NY, USA: 2010. Pseudoknot-dependent programmed −1 ribosomal frameshifting: Structures, mechanisms and models; pp. 149–174.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

