RNA-Binding Proteins as Regulators of Internal Initiation of Viral mRNA Translation
- PMID: 35215780
- PMCID: PMC8879377
- DOI: 10.3390/v14020188
RNA-Binding Proteins as Regulators of Internal Initiation of Viral mRNA Translation
Abstract
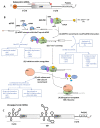
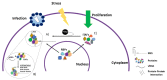
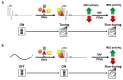
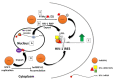
Viruses are obligate intracellular parasites that depend on the host's protein synthesis machinery for translating their mRNAs. The viral mRNA (vRNA) competes with the host mRNA to recruit the translational machinery, including ribosomes, tRNAs, and the limited eukaryotic translation initiation factor (eIFs) pool. Many viruses utilize non-canonical strategies such as targeting host eIFs and RNA elements known as internal ribosome entry sites (IRESs) to reprogram cellular gene expression, ensuring preferential translation of vRNAs. In this review, we discuss vRNA IRES-mediated translation initiation, highlighting the role of RNA-binding proteins (RBPs), other than the canonical translation initiation factors, in regulating their activity.
Keywords: IRES; IRES-transacting factor; ITAF; RBP; RNA-binding protein; internal ribosome entry site.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Abrahao J., Silva L., Silva L.S., Khalil J.Y.B., Rodrigues R., Arantes T., Assis F., Boratto P., Andrade M., Kroon E.G., et al. Tailed giant Tupanvirus possesses the most complete translational apparatus of the known virosphere. Nat. Commun. 2018;9:749. doi: 10.1038/s41467-018-03168-1. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

