SARS-CoV-2: Ultrastructural Characterization of Morphogenesis in an In Vitro System
- PMID: 35215794
- PMCID: PMC8879486
- DOI: 10.3390/v14020201
SARS-CoV-2: Ultrastructural Characterization of Morphogenesis in an In Vitro System
Abstract
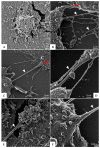
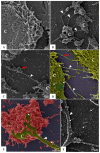
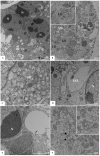
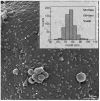
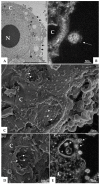
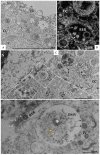
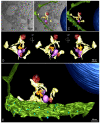
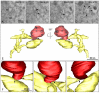
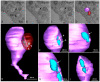
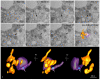
The pandemic caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has impacted public health and the world economy and fueled a worldwide race to approve therapeutic and prophylactic agents, but so far there are no specific antiviral drugs. Understanding the biology of the virus is the first step in structuring strategies to combat it, and in this context several studies have been conducted with the aim of understanding the replication mechanism of SARS-CoV-2 in vitro systems. In this work, studies using transmission and scanning electron microscopy and 3D electron microscopy modeling were performed with the goal of characterizing the morphogenesis of SARS-CoV-2 in Vero-E6 cells. Several ultrastructural changes were observed-such as syncytia formation, cytoplasmic membrane projections, lipid droplets accumulation, proliferation of double-membrane vesicles derived from the rough endoplasmic reticulum, and alteration of mitochondria. The entry of the virus into cells occurred through endocytosis. Viral particles were observed attached to the cell membrane and in various cellular compartments, and extrusion of viral progeny took place by exocytosis. These findings allow us to infer that Vero-E6 cells are highly susceptible to SARS-CoV-2 infection as described in the literature and their replication cycle is similar to that described with SARS-CoV and MERS-CoV in vitro models.
Keywords: 3D electron microscopy modeling; SARS-CoV-2; Vero-E6 cells; morphogenesis; scanning electron microscopy; transmission electron microscopy; ultrastructural studies.
Conflict of interest statement
The authors declare no conflict of interest exists.
Figures



References
-
- Gorbalenya A.E., Baker S.C., Baric R.S., Groot R.J., Drosten C., Gulyaeva A.A., Haagmans B.L., Lauber C., Leontovich A.M., Neuman B.W., et al. The species severe acute respiratory syndrome-related coronavirus: Classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5:536–544. - PMC - PubMed
-
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K.W., Bleicker T., Brünink S., Schneider J., Schmidt M.L., et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance. 2020;25:2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

