ViralFlow: A Versatile Automated Workflow for SARS-CoV-2 Genome Assembly, Lineage Assignment, Mutations and Intrahost Variant Detection
- PMID: 35215811
- PMCID: PMC8877152
- DOI: 10.3390/v14020217
ViralFlow: A Versatile Automated Workflow for SARS-CoV-2 Genome Assembly, Lineage Assignment, Mutations and Intrahost Variant Detection
Abstract
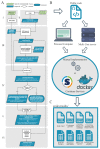
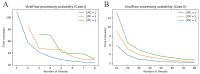
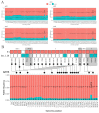
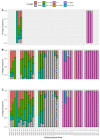
The COVID-19 pandemic is driven by Severe Acute Respiratory Syndrome coronavirus 2 (SARS-CoV-2) that emerged in 2019 and quickly spread worldwide. Genomic surveillance has become the gold standard methodology used to monitor and study this fast-spreading virus and its constantly emerging lineages. The current deluge of SARS-CoV-2 genomic data generated worldwide has put additional pressure on the urgent need for streamlined bioinformatics workflows. Here, we describe a workflow developed by our group to process and analyze large-scale SARS-CoV-2 Illumina amplicon sequencing data. This workflow automates all steps of SARS-CoV-2 reference-based genomic analysis: data processing, genome assembly, PANGO lineage assignment, mutation analysis and the screening of intrahost variants. The pipeline is capable of processing a batch of around 100 samples in less than half an hour on a personal laptop or in less than five minutes on a server with 50 threads. The workflow presented here is available through Docker or Singularity images, allowing for implementation on laptops for small-scale analyses or on high processing capacity servers or clusters. Moreover, the low requirements for memory and CPU cores and the standardized results provided by ViralFlow highlight it as a versatile tool for SARS-CoV-2 genomic analysis.
Keywords: SARS-CoV-2; genomic variants; genomics; genotyping; software; virus bioinformatics; viruses.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- WHO Director-General’s Opening Remarks at the Media Briefing on COVID-19—11 March 2020. [(accessed on 27 September 2021)]. Available online: https://www.who.int/director-general/speeches/detail/who-director-genera....
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

