HIV-1 Nucleocapsid Protein Binds Double-Stranded DNA in Multiple Modes to Regulate Compaction and Capsid Uncoating
- PMID: 35215829
- PMCID: PMC8879225
- DOI: 10.3390/v14020235
HIV-1 Nucleocapsid Protein Binds Double-Stranded DNA in Multiple Modes to Regulate Compaction and Capsid Uncoating
Abstract
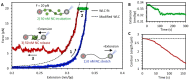
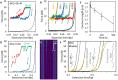
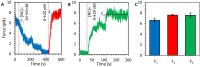
The HIV-1 nucleocapsid protein (NC) is a multi-functional protein necessary for viral replication. Recent studies have demonstrated reverse transcription occurs inside the fully intact viral capsid and that the timing of reverse transcription and uncoating are correlated. How a nearly 10 kbp viral DNA genome is stably contained within a narrow capsid with diameter similar to the persistence length of double-stranded (ds) DNA, and the role of NC in this process, are not well understood. In this study, we use optical tweezers, fluorescence imaging, and atomic force microscopy to observe NC binding a single long DNA substrate in multiple modes. We find that NC binds and saturates the DNA substrate in a non-specific binding mode that triggers uniform DNA self-attraction, condensing the DNA into a tight globule at a constant force up to 10 pN. When NC is removed from solution, the globule dissipates over time, but specifically-bound NC maintains long-range DNA looping that is less compact but highly stable. Both binding modes are additionally observed using AFM imaging. These results suggest multiple binding modes of NC compact DNA into a conformation compatible with reverse transcription, regulating the genomic pressure on the capsid and preventing premature uncoating.
Keywords: DNA condensation; HIV-1 nucleocapsid protein; atomic force microscopy; capsid uncoating; optical tweezers.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures



Similar articles
-
Cationic Residues of the HIV-1 Nucleocapsid Protein Enable DNA Condensation to Maintain Viral Core Particle Stability during Reverse Transcription.Viruses. 2024 May 29;16(6):872. doi: 10.3390/v16060872. Viruses. 2024. PMID: 38932164 Free PMC article.
-
Reverse Transcription Mechanically Initiates HIV-1 Capsid Disassembly.J Virol. 2017 May 26;91(12):e00289-17. doi: 10.1128/JVI.00289-17. Print 2017 Jun 15. J Virol. 2017. PMID: 28381579 Free PMC article.
-
The HIV-1 nucleocapsid chaperone protein forms locally compacted globules on long double-stranded DNA.Nucleic Acids Res. 2021 May 7;49(8):4550-4563. doi: 10.1093/nar/gkab236. Nucleic Acids Res. 2021. PMID: 33872352 Free PMC article.
-
May I Help You with Your Coat? HIV-1 Capsid Uncoating and Reverse Transcription.Int J Mol Sci. 2024 Jun 28;25(13):7167. doi: 10.3390/ijms25137167. Int J Mol Sci. 2024. PMID: 39000271 Free PMC article. Review.
-
Overview of the Nucleic-Acid Binding Properties of the HIV-1 Nucleocapsid Protein in Its Different Maturation States.Viruses. 2020 Sep 29;12(10):1109. doi: 10.3390/v12101109. Viruses. 2020. PMID: 33003650 Free PMC article. Review.
Cited by
-
SARS-CoV-2 Nucleocapsid Protein Has DNA-Melting and Strand-Annealing Activities With Different Properties From SARS-CoV-2 Nsp13.Front Microbiol. 2022 Jul 22;13:851202. doi: 10.3389/fmicb.2022.851202. eCollection 2022. Front Microbiol. 2022. PMID: 35935242 Free PMC article.
-
Protamine folds DNA into flowers and loop stacks.Biophys J. 2023 Nov 7;122(21):4288-4302. doi: 10.1016/j.bpj.2023.10.003. Epub 2023 Oct 6. Biophys J. 2023. PMID: 37803830 Free PMC article.
-
Cationic Residues of the HIV-1 Nucleocapsid Protein Enable DNA Condensation to Maintain Viral Core Particle Stability during Reverse Transcription.Viruses. 2024 May 29;16(6):872. doi: 10.3390/v16060872. Viruses. 2024. PMID: 38932164 Free PMC article.
-
Structural domains of SARS-CoV-2 nucleocapsid protein coordinate to compact long nucleic acid substrates.Nucleic Acids Res. 2023 Jan 11;51(1):290-303. doi: 10.1093/nar/gkac1179. Nucleic Acids Res. 2023. PMID: 36533523 Free PMC article.
-
HIV-1 uncoating requires long double-stranded reverse transcription products.Sci Adv. 2024 Apr 26;10(17):eadn7033. doi: 10.1126/sciadv.adn7033. Epub 2024 Apr 24. Sci Adv. 2024. PMID: 38657061 Free PMC article.
References
-
- Coffin J.M., Hughes S.H., Varmus H.E. Retroviruses. [(accessed on 31 August 2021)];1997 Available online: https://www.ncbi.nlm.nih.gov/books/NBK19376/
-
- Levin J.G., Guo J., Rouzina I., Musier-Forsyth K. Nucleic acid chaperone activity of HIV-1 nucleocapsid protein: Critical role in reverse transcription and molecular mechanism. Prog. Nucleic Acid Res. Mol. Biol. 2005;80:217–286. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

