Epidemiology and Genetic Analysis of SARS-CoV-2 in Myanmar during the Community Outbreaks in 2020
- PMID: 35215852
- PMCID: PMC8875553
- DOI: 10.3390/v14020259
Epidemiology and Genetic Analysis of SARS-CoV-2 in Myanmar during the Community Outbreaks in 2020
Abstract
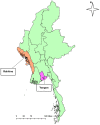
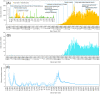
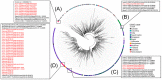
We aimed to analyze the situation of the first two epidemic waves in Myanmar using the publicly available daily situation of COVID-19 and whole-genome sequencing data of SARS-CoV-2. From March 23 to December 31, 2020, there were 33,917 confirmed cases and 741 deaths in Myanmar (case fatality rate of 2.18%). The first wave in Myanmar from March to July was linked to overseas travel, and then a second wave started from Rakhine State, a western border state, leading to the second wave spreading countrywide in Myanmar from August to December 2020. The estimated effective reproductive number (Rt) nationwide reached 6-8 at the beginning of each wave and gradually decreased as the epidemic spread to the community. The whole-genome analysis of 10 Myanmar SARS-CoV-2 strains together with 31 previously registered strains showed that the first wave was caused by GISAID clade O or PANGOLIN lineage B.6 and the second wave was changed to clade GH or lineage B.1.36.16 with a close genetic relationship with other South Asian strains. Constant monitoring of epidemiological situations combined with SARS-CoV-2 genome analysis is important for adjusting public health measures to mitigate the community transmissions of COVID-19.
Keywords: COVID-19; SARS-CoV-2; case fatality rate; community transmission; molecular epidemiology; reproductive number; whole-genome sequences.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures

References
-
- World Health Organization Coronavirus Disease 2019 (COVID-19). Situation Report—51. [(accessed on 30 May 2020)]. Available online: https://apps.who.int/iris/bitstream/handle/10665/331475/nCoVsitrep11Mar2....
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

