Characterization of the Novel Phage vB_VpaP_FE11 and Its Potential Role in Controlling Vibrio parahaemolyticus Biofilms
- PMID: 35215857
- PMCID: PMC8879856
- DOI: 10.3390/v14020264
Characterization of the Novel Phage vB_VpaP_FE11 and Its Potential Role in Controlling Vibrio parahaemolyticus Biofilms
Abstract
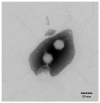
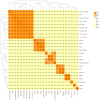
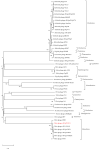
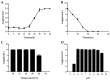
Vibrio parahaemolyticus causes aquatic vibriosis. Its biofilm protects it from antibiotics; therefore, a new different method is needed to control V. parahaemolyticus for food safety. Phage therapy represents an alternative strategy to control biofilms. In this study, the lytic Vibrio phage vB_VpaP_FE11 (FE11) was isolated from the sewers of Guangzhou Huangsha Aquatic Market. Electron microscopy analysis revealed that FE11 has a typical podovirus morphology. Its optimal stability temperature and pH range were found to be 20-50 °C and 5-10 °C, respectively. It was completely inactivated following ultraviolet irradiation for 20 min. Its latent period is 10 min and burst size is 37 plaque forming units/cell. Its double-stranded DNA genome is 43,397 bp long, with a G + C content of 49.24% and 50 predicted protein-coding genes. As a lytic phage, FE11 not only prevented the formation of biofilms but also could destroy the formed biofilms effectively. Overall, phage vB_VpaP_FE11 is a potential biological control agent against V. parahaemolyticus and the biofilm it produces.
Keywords: Vibrio parahaemolyticus; aquaculture; biofilm; biological control; phage.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as potential conflicts of interest.
Figures



Similar articles
-
Characterization of novel bacteriophages for effective phage therapy against Vibrio infections in aquaculture.J Microbiol. 2025 May;63(5):e2502009. doi: 10.71150/jm.2502009. Epub 2025 May 27. J Microbiol. 2025. PMID: 40468661
-
A virulent phage vB_VpaP_R28Z infecting Vibrio parahaemolyticus with potential for therapeutic application.BMC Microbiol. 2025 Jul 12;25(1):433. doi: 10.1186/s12866-025-04133-x. BMC Microbiol. 2025. PMID: 40646434 Free PMC article.
-
Genome characterization of the novel lytic Vibrio parahaemolyticus phage vB_VpP_BA6.Arch Virol. 2019 Oct;164(10):2627-2630. doi: 10.1007/s00705-019-04351-5. Epub 2019 Jul 30. Arch Virol. 2019. PMID: 31363923
-
Targeted phytogenic compounds against Vibrio parahaemolyticus biofilms.Crit Rev Food Sci Nutr. 2025;65(9):1761-1772. doi: 10.1080/10408398.2023.2299949. Epub 2024 Jan 8. Crit Rev Food Sci Nutr. 2025. PMID: 38189321 Review.
-
Bacteriophage therapy in aquaculture: current status and future challenges.Folia Microbiol (Praha). 2022 Aug;67(4):573-590. doi: 10.1007/s12223-022-00965-6. Epub 2022 Mar 19. Folia Microbiol (Praha). 2022. PMID: 35305247 Review.
Cited by
-
Characteristics and whole-genome analysis of a novel Pseudomonas syringae pv. tomato bacteriophage D6 isolated from a karst cave.Virus Genes. 2024 Jun;60(3):295-308. doi: 10.1007/s11262-024-02064-9. Epub 2024 Apr 9. Virus Genes. 2024. PMID: 38594490 Free PMC article.
-
Characterization of the novel broad-spectrum lytic phage Phage_Pae01 and its antibiofilm efficacy against Pseudomonas aeruginosa.Front Microbiol. 2024 Jul 17;15:1386830. doi: 10.3389/fmicb.2024.1386830. eCollection 2024. Front Microbiol. 2024. PMID: 39091310 Free PMC article.
-
Characterization and antimicrobial activity of a novel lytic phage vB_SmaS_QH16 against Stenotrophomonas maltophilia: in vitro, in vivo, and biofilm studies.Front Cell Infect Microbiol. 2025 Jul 10;15:1610857. doi: 10.3389/fcimb.2025.1610857. eCollection 2025. Front Cell Infect Microbiol. 2025. PMID: 40708751 Free PMC article.
-
Characterization and Genome Analysis of Vibrio campbellii Lytic Bacteriophage OPA17.Microbiol Spectr. 2023 Jan 31;11(2):e0162322. doi: 10.1128/spectrum.01623-22. Online ahead of print. Microbiol Spectr. 2023. PMID: 36719217 Free PMC article.
-
A novel virus potentially evolved from the N4-like viruses represents a unique viral family: Poorviridae.Appl Environ Microbiol. 2024 Dec 18;90(12):e0155924. doi: 10.1128/aem.01559-24. Epub 2024 Nov 21. Appl Environ Microbiol. 2024. PMID: 39570022 Free PMC article.
References
-
- Hong X., Lu L., Xu D. Progress in Research on Acute Hepatopancreatic Necrosis Disease (AHPND) Aquacult. Int. 2016;24:577–593. doi: 10.1007/s10499-015-9948-x. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

