Discovery and Evolutionary Analysis of a Novel Bat-Borne Paramyxovirus
- PMID: 35215881
- PMCID: PMC8879077
- DOI: 10.3390/v14020288
Discovery and Evolutionary Analysis of a Novel Bat-Borne Paramyxovirus
Abstract
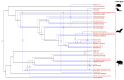
Paramyxoviruses are a group of RNA viruses, such as mumps virus, measles virus, Nipah virus, Hendra virus, Newcastle disease virus, and parainfluenza virus, usually transmitted by airborne droplets that are predominantly responsible for acute respiratory diseases. In this paper, we identified a novel paramyxovirus belonging to genus Jeilongvirus infecting 4/112 (3.6%) bats from two trapping sites of Hainan Province of China. In these animals, the viral RNA was detected exclusively in kidney tissues. This is the first full-length Jeilongvirus genome (18,095 nucleotides) from bats of genus Hipposideros, which exhibits a canonical genome organization and encodes SH and TM proteins. Results, based on phylogenic analysis and genetic distances, indicate that the novel paramyxovirus formed an independent lineage belonging to genus Jeilongvirus, representing, thus, a novel species. In addition, the virus-host macro-evolutionary analysis revealed that host-switching was not only a common co-phylogenetic event, but also a potential mechanism by which rats are infected by bat-origin Jeilongvirus through cross-species virus transmission, indicating a bat origin of the genus Jeilongvirus. Overall, our study broadens the viral diversity, geographical distribution, host range, and evolution of genus Jeilongvirus.
Keywords: Jeilongvirus; bat; co-evolutionary; paramyxovirus; virus discovery.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Vanmechelen B., Bletsa M., Laenen L., Lopes A.R., Vergote V., Beller L., Deboutte W., Korva M., Avšič Županc T., Goüy de Bellocq J., et al. Discovery and genome characterization of three new Jeilongviruses, a lineage of paramyxoviruses characterized by their unique membrane proteins. BMC Genom. 2018;19:617. doi: 10.1186/s12864-018-4995-0. - DOI - PMC - PubMed
-
- Lee S.H., Kim K., Kim J., No J.S., Park K., Budhathoki S., Lee S.H., Lee J., Cho S.H., Cho S., et al. Discovery and Genetic Characterization of Novel Paramyxoviruses Related to the Genus Henipavirus in Crocidura Species in the Republic of Korea. Viruses. 2021;13:2020. doi: 10.3390/v13102020. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

