Evidence of a Possible Viral Host Switch Event in an Avipoxvirus Isolated from an Endangered Northern Royal Albatross (Diomedea sanfordi)
- PMID: 35215898
- PMCID: PMC8880153
- DOI: 10.3390/v14020302
Evidence of a Possible Viral Host Switch Event in an Avipoxvirus Isolated from an Endangered Northern Royal Albatross (Diomedea sanfordi)
Abstract
Avipoxviruses have been characterized from many avian species. Two recent studies have reported avipoxvirus-like viruses with varying pathogenicity in reptiles. Avipoxviruses are considered to be restricted to avian hosts. However, reports of avipoxvirus-like viruses from reptiles such as the green sea turtle (Chelonia mydas) and crocodile tegu (Crocodilurus amazonicus) suggest that cross-species transmission, within avian species and beyond, may be possible. Here we report evidence for a possible host switching event with a fowlpox-like virus recovered from an endangered northern royal albatross (Diomodea sanfordi)-a species of Procellariiformes, unrelated to Galliformes, not previously known to have been infected with fowlpox-like viruses. Complete genome sequencing of this virus, tentatively designated albatrosspox virus 2 (ALPV2), contained many fowlpox virus-like genes, but also 63 unique genes that are not reported in any other poxvirus. The ALPV2 genome contained 296 predicted genes homologous to different avipoxviruses, 260 of which were homologous to an American strain of fowlpox virus (FWPV). Subsequent phylogenetic analyses indicate that ALPV2 likely originated from a fowlpox virus-like progenitor. These findings highlight the importance of host-switching events where viruses cross species barriers with the risk of disease in close and distantly related host populations.
Keywords: avipoxvirus; evolution; host-switch; northern royal albatross.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

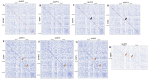

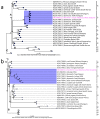
References
-
- Croxall J.P., Butchart S.H.M., Lascelles B.E.N., Stattersfield A.J., Sullivan B.E.N., Symes A., Taylor P. Seabird conservation status, threats and priority actions: A global assessment. Bird Conserv. Int. 2012;22:1–34. doi: 10.1017/S0959270912000020. - DOI
-
- Croxall J.P., Gales R. An Assessment of the Conservation Status of Albatrosses. In: Robertson G., Gales R., editors. Albatross Biology and Conservation. Surrey Beatty & Sons; Chipping Norton, UK: 1998. pp. 46–65.
-
- Cooper J., Baker G.B., Double M.C., Gales R., Papworth W., Tasker M.L., Waugh S.M. The agreement on the conservation of albatrosses and petrels: Rationale, history, progress and the way forward. Mar. Ornithol. 2006;34:1–5.
-
- Marcela M., Uhart L.G., Flavio Q. Review of diseases (pathogen isolation, direct recovery and antibodies) in albatrosses and large petrels worldwide. Bird Conserv. Int. 2018;28:169–196.
-
- BirdLife I. Diomedea sanfordi. The IUCN Red List of Threatened Species 2018: E.T22728323A132656392. [(accessed on 30 January 2021)]. Available online: - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

