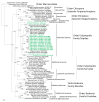
Figure 2
Phylogenetic trees generated by the Bayesian method, under the best-fit GTR+I+Γ model of evolution, based on the S-, M-, and L-genomic segments of ACDV (green, bold lettering). The phylogenetic positions of ACDV strains in Russia are shown in relationship to mole-borne hantaviruses (green): Asama orthohantavirus (ASAV N10, S: EU929072; M: EU929075; L: EU929078) from Urotrichus talpoides, Nova mobatvirus (NVAV Te34, S: KR072621, M: KR072622, L: KR072623), and Bruges orthohantavirus (BRGV BE/VieuxGenappeTE2013, S: KR072621; M: KR072622; L: KR072623 and BRGV DEWandlitzTE2013, S: MF683844; M: MF683845; L: MF683846) from Talpa europaea, Rockport orthohantavirus (RKPV MSB57412, S: HM015223; M: HM015222; L: HM015221) from Scalopus aquaticus, and Oxbow orthohantavirus (OXBV Ng1453, S: FJ5339166; M: FJ539167; L: FJ593497) from Neurotrichus gibbsii. Shrew-borne hantaviruses (red lettering) include Thottapalayam thottimvirus (TPMV VRC66412, S: AY526097, M: EU001329, L: EU001330) from Suncus murinus, Imjin thottimvirus (MJNV Cl05-11, S: EF641804; M: EF641798; L: EF641806) from Crocidura lasiura, Uluguru thottimvirus (ULUV FMNH158302, S: JX193695; M: JX193696; L: JX193697) from Myosorex geata, Kilimanjaro thottimvirus (KMJV FMNH174124, S: JX193698; M: JX193699; L: JX193700) from Myosorex zinki, Jeju orthohantavirus (JJUV SH42, S: HQ663933; M: HQ663934; L: HQ663935) from Crocidura shantungensis, Cao Bằng orthohantavirus (CBNV TC-3, S: EF543524; M: EF543526; L: EF543525) from Anourosorex squamipes, Azagny orthohantavirus (AZGV KBM15, S: JF276226; M: JF276227; L: JF276228) from Crocidura obscurior, Bowé orthohantavirus (BOWV VN1512, S: KC631782; M: KC631783; L: KC631784) from Crocidura douceti, prototype Seewis orthohantavirus (SWSV mp70, S: EF636024; M: EF636025; L: EF636026), and Altai virus (ALTV ALT302, S: MK340902; M: MK340903; L: MT648514) from Sorex araneus, Lena virus (LENV Khekhtsir-Sc67, S: MH499470; M: MH499471; L: MH499472) from Sorex caecutiens, Asikkala virus (ASIV CZ/Drahany/420/2010/Sm, S: KC880342; M: KC880345; L: KC880348) from Sorex minutus, Jemez Springs orthohantavirus (JMSV MSB144475, S: FJ593499; M: FJ593500; L: FJ593501) from Sorex monticolus, Tanganya virus (TGNV Tan826, S: EF050455; L: EF050454) from Crocidura theresea, Ash River virus (ARRV MSB73418, S: EF650086; L: EF619961) from Sorex cinereus, Yakeshi virus (YKSV Si-210, S: JX465423; M: JX465403; L: JX465389) from Sorex isodon, Kenkeme virus (KKMV MSB148794, S: GQ306148; M: GQ306149; L: GQ306150 and KKMV Fuyuan Sr326, S: NC_034559; M: KJ857337; L: KJ857320) from Sorex roboratus, and Boginia virus (BOGV 3486, L: KM394262) from Neomys fodiens. Also shown are representative rodent-borne hantaviruses (black lettering), including Sin Nombre orthohantavirus (SNV NMH10, S: NC_005216; M: NC_005215; L: NC_005217), Andes orthohantavirus (ANDV Chile9717869, S: AF291702; M: AF291703; L: AF291704), Prospect Hill orthohantavirus (PHV PH-1, S: Z49098; M: X55129; L: EF646763), Tula orthohantavirus (TULV M5302v, S: NC_005227; M: NC_005228; L: NC_005226), Puumala orthohantavirus (PUUV Sotkamo, S: NC_005224; M: NC_005223; L: NC_005225), Sangassou orthohantavirus (SANGV SA14, S: JQ082300; M: JQ082301; L: JQ082302), Soochong orthohantavirus (SOOV SOO-1, S: AY675349; M: AY675353; L: DQ056292), Dobrava/Belgrade orthohantavirus (DOBV/BGDV Greece, S: NC_005233; M: NC_005234; L: NC_005235), Hantaan orthohantavirus (HTNV 76-118, S: NC_005218; M: NC_005219; L: NC_005222), and Seoul orthohantavirus (SEOV 80-39, S: NC_005236; M: NC_005237; L: NC_005238), and Tigray virus (TIGV ET2121, S: KU934010; M: KU934009; L: KU934008) from Stenocephalemys albipes. Bat-borne hantaviruses (blue lettering) include Brno loanvirus (BRNV 7/2012/CZE, S: KX845678; M: KX845679; L: KX845680) from Nyctalus noctula, Láibīn mobatvirus (LAIV BT20, S: KM102247; M: KM102248; L: KM102249) from Taphozous melanopogon, Xuân Sơn mobatvirus (XSV VN1982B4, S: KC688335; L: JX912953) from Hipposideros pomona, Quezon mobatvirus (QZNV MT1720/1657, S: KU950713; M: KU950714; L: KU950715) from Rousettus amplexicaudatus, Mouyassué virus (MOYV KB576, L: JQ28771) from Neoromicia nanus, Makokou virus (MAKV GB303, L: KT316176) from Hipposideros ruber, Huángpí virus (HUPV Pa-1, S: JX473273, and L: JX465369) from Pipistrellus abramus, Lóngquán loanvirus (LQUV Ra-25, S: JX465415; M: JX465397; and L: JX465381) from Rhinolophus sinicus, Đakrông mobatvirus (DKGV VN2913B72, S: MG663536; M: MG663535; L: MG663534) from Aselliscus stoliczkanus, respectively. The numbers at each node are posterior node probabilities (>0.7), based on 150,000 trees: two replicate Markov Chain Monte Carlo runs consisting of six chains of 10 million generations each sampled every 100 generations with a burn-in of 25,000 (25%). The scale bar indicates the nucleotide substitutions per site.